Family: Geminiviridae
Genus: Becurtovirus
Distinguishing features
Although the biological properties of known becurtoviruses are similar to those of curtoviruses, they share only ~60% genome-wide sequence identity. The unique feature of the becurtovirus genome is that the coat protein (CP) is curtovirus-like, whereas the replication-associated protein (Rep) is most closely related to those of mastreviruses. Additionally, becurtoviruses (and eragroviruses) have a unique nonanucleotide at the v-ori that differs from those of all other geminiviruses (Yazdi et al., 2008).
Virion
See discussion under family description.
Genome organization and replication
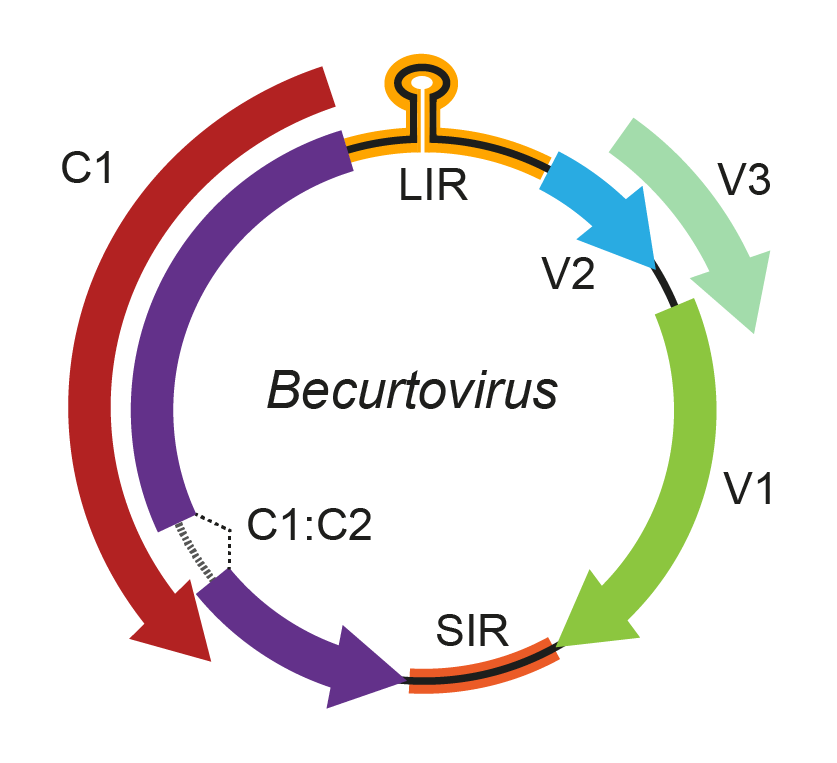
The becurtovirus genome comprises three overlapping ORFs (V1, V2, and V3) in the virion-sense strand and two complementary-sense ORFs (C1 and C2) (Figure 1. Becurtovirus) (Varsani et al., 2014b). The V1, V2 and V3 ORFs are analogous to their curtovirus counterparts, encoding, respectively, the coat protein, movement protein and a protein involved in regulating the ss/ds DNA ratio. The C1 and C2 ORFs, however, are more related to the mastrevirus ORFs occupying the same position in the genome. Furthermore, in contrast to curtoviruses, becurtoviruses lack C3 and C4 ORFs (Yazdi et al., 2008). Based on the evidence from mastreviruses, it is likely that the Rep ORF is expressed from spliced transcripts, and contains an intron sequence. As is the case with the eragroviruses, becurtoviruses have a 5′-TAAGATTCC-3′ nonanucleotide sequence at the v-ori.
|
|
|
Figure 1. .Becurtovirus. Genomic organization of members of the genus Betacurtovirus. ORFs are denoted as being encoded on the virion-sense (V) or complementary-sense (C) strand. The position of the stem-loop containing the conserved 5′-TAAGATTCC-3′ sequence located in the long intergenic region (LIR) is shown. An intron is predicted to occur between ORFs C1 and C2. SIR, short intergenic region. |
Biology
Host range
Isolates of beet curly top Iran virus (BCTIV) have been obtained from the dicotyledonous species Beta vulgaris subsp. vulgaris (beet), Beta vulgaris subsp. maritima (sea beet), Vigna unguiculata (cowpea), Solanum lycopersicum (tomato) and Phaseolus vulgaris (common bean) (Razavinejad et al., 2013). Isolates of spinach curly top Arizona virus (SCTAV) have been found infecting the dicot species Spinacia oleracea (spinach) (Hernandez-Zepeda et al., 2013) and the only isolate available from Exomis microphylla latent virushas been obtained from Exomis microphylla, a wild plant of the family Amaranthaceae.
Transmission
The leafhopper Circulifer haematoceps was shown to transmit beet curly top Iran virus to sugar beet under greenhouse conditions, resulting in typical curly top symptoms (Heydarnejad et al., 2013).
Species demarcation criteria
Nucleotide sequence identity: Based on the distribution of pairwise identities, a species demarcation threshold of 80% is currently used (Varsani et al., 2014b). Thus, pairs of becurtovirus full genome sequences with >80% pairwise identity are considered as members of the same species. This criterion is tentative in that it is based on information available on member of only three species. As more becurtovirus sequences are discovered, this threshold may change.