Family: Geminiviridae
Genus: Grablovirus
Distinguishing features
The monopartite genome of grabloviruses is about 3.2 kb, significantly longer than those of other monopartite geminiviruses, which range from 2.7 to 3.0 kb. Its genomic organization is unlike that of other geminiviruses, with virion and complementary sense ORFs that have no homologues in members of the other genera in the family.
Virion
See discussion under family description.
Genome organization and replication
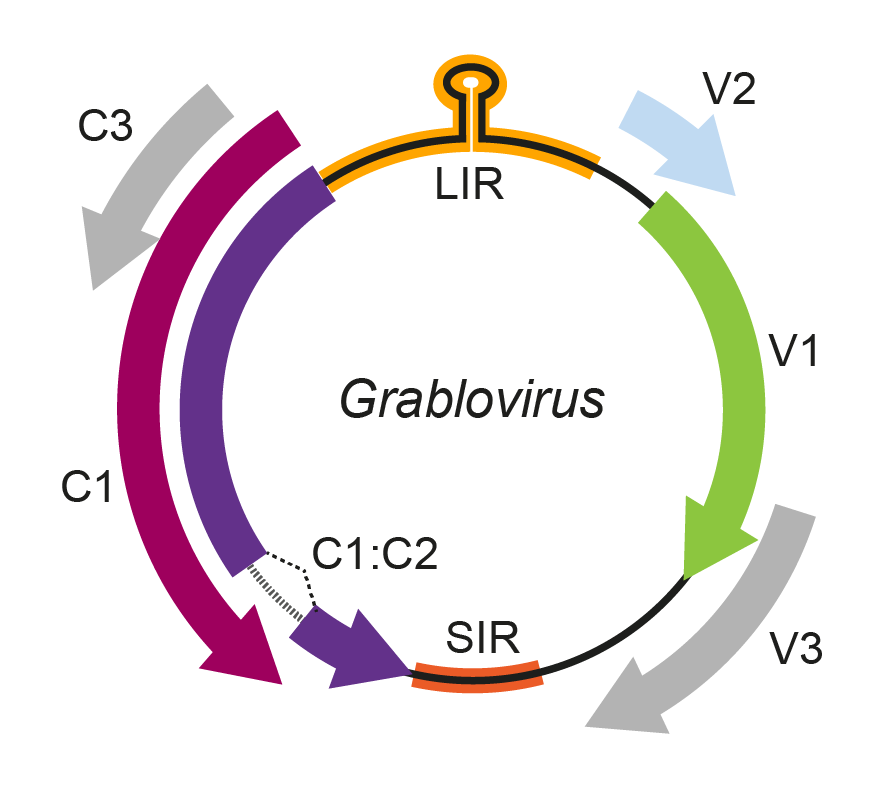
The grablovirus genome has three ORFs in the virion sense strand (V1, V2, and V3) and three in the complementary sense strand (C1, C2, and C3) (Figure 1. Grablovirus). By analogy with mastreviruses, the C1 and C2 ORFs likely encode the replication-associated protein (Rep) from a spliced transcript, and RepA is encoded from the unspliced C1 transcript (Krenz et al., 2014). C3 is entirely embedded in the C1 ORF in the same reading frame. Based on its position in the genome and sequence identity with other geminiviruses, the V1 ORF is predicted to encode the grablovirus coat protein. Likewise, by analogy with other geminiviruses, the V2 and V3 ORFs may encode movement proteins (Sudarshana et al., 2015). Grabloviruses have the v-ori nonanucleotide motif 5′-TAATATTAC-3′.
|
|
|
Figure 1. Grablovirus. Genomic organization of grabloviruses. ORFs are denoted as being encoded on the virion-sense (V) or complementary-sense (C) strand,. The position of the stem-loop containing the conserved 5′-TAATATTAC-3′ sequence located in the long intergenic region (LIR) is shown. An intron is predicted to occur between ORFs C1 and C2. SIR, short intergenic region. |
Biology
Host range
Grapevine red blotch virus is known to infect only grapevine. Prunus latent virus infects stone fruit trees of the genus Prunus (apricot, plum and cherry) and wild Vitis latent virus a non-cultivated grapevine (Vitis sp.)
Transmission
The three-cornered alfalfa treehopper, Spissistilus festinus (Hemiptera: Membracidae), is capable of both acquiring the virus from infected grapevines and transmitting the virus to grapevines in the laboratory (Bahder et al., 2016).
Species demarcation criteria
Nucleotide sequence identity: isolates sharing <80% genome-wide pairwise identity with members of grablovirus species should be classified as members of a distinct species. This is a tentative species demarcation criterion that will need to be reassessed as additional grablovirus sequences become available.