Family: Geminiviridae
Genus: Curtovirus
Distinguishing features
Curtoviruses are monopartite geminiviruses that are vectored by leafhoppers, often in a species-specific manner. In common with viruses in the genus Begomovirus, curtoviruses occur in both the New and Old World. They infect a wide variety of vegetable crops and are thought to be widely prevalent in non-cultivated dicot species (Varsani et al., 2014a). Curtovirus genomes usually have seven genes, but isolates have been discovered that have only five or six genes. Curtovirus genomes may be associated with defective-interfering DNAs (DI particles) that function in some instances to reduce symptom severity.
Virion
See discussion under family description.
Genome organization and replication
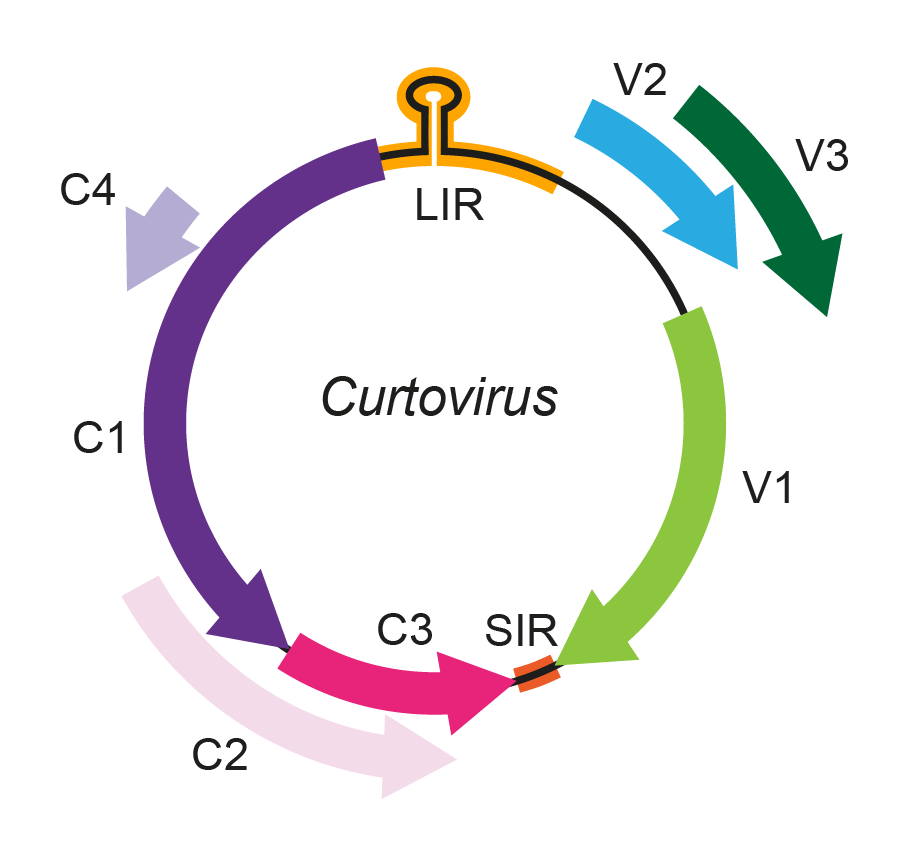
The genome of curtoviruses consists of a single circular ssDNA component of 2.9–3.0 kb, encoding six to seven proteins (Figure 1. Curtovirus). The three ORFs encoded on the virion-sense strand are the coat protein (CP, ORF V1), which encapsidates the virion-sense ssDNA and is involved in virus movement and insect vector transmission, a movement protein (MP, ORF V2), and V3, which is involved in the regulation of the relative levels of ssDNA and dsDNA. The complementary-sense strand encodes the replication-associated protein (Rep, ORF C1), which is required for the initiation of viral DNA replication, the C2 protein (ORF C2), which acts as a pathogenicity factor in some hosts, a replication enhancer protein (REn, ORF C3), and the C4 protein (ORF C4), which is an important symptom determinant implicated in cell-cycle control (Hanley-Bowdoin et al., 2013). Nucleotide sequence comparisons suggest that curtoviruses and begomoviruses diverged after a recombination event altered insect vector specificity (Rybicki 1994).
|
|
|
Figure 1. Curtovirus. Genomic organization of curtoviruses. ORFs are denoted as being encoded on the virion-sense (V) or complementary-sense (C) strand, and corresponding protein products are indicated where these are known. ORF C3 is not present in horseradish curly top virus (HrCTV). The position of the stem-loop containing the conserved 5′-TAATATTAC-3′ sequence located in the long intergenic region (LIR) is shown. CP, coat protein; MP, movement protein; Rep, replication-associated protein; REn, replication enhancer protein; TrAP, transcriptional activator protein. |
Biology
Host range
Isolates of the type species, Beet curly top virus, have a very wide host range within dicot plants, including over 300 species in 44 plant families (Strausbaugh et al., 2008).
Transmission
Curtoviruses are transmitted by specific leafhoppers (order Hemiptera, family Cicadellidae) in a persistent (circulative, non-propagative) manner (Soto and Gilbertson 2003). BCTV may be transmitted with difficulty by mechanical inoculation. Most members are transmitted experimentally to plants by Agrobacterium-mediated transfer (agroinoculation) from partially or tandemly repeated cloned genomic DNA.
Antigenicty
Serological tests show beet curly top virus (BCTV), tomato leaf roll virus (TLRV) and tomato pseudo-curly top virus (TPCTV, genus Topocuvirus) to be relatively closely related. Distant serological relationships are observed between curtoviruses and begomoviruses (Harrison and Robinson 1999).
Species demarcation criteria
The following criteria should be used as a guideline to establish taxonomic status:
- Nucleotide sequence identity: Pairs of genomes with <77% pairwise identity calculated by pairwise alignment ignoring gapped sites should be considered members of the same species (Varsani et al., 2014a), even in the absence of additional data. Nonetheless, when approaching the cut-off value, decisions based sequence comparisons should take into account the biological properties of the virus.
- Trans-replication of genomic components: the inability of the Rep protein of one virus to trans-replicate the genomic component of a second virus may be used as an indicator that the two viruses belong to different species.
- Coat protein serology: CP antigenicity may differ between members of different species, although the high level of sequence conservation of this protein suggests that this criterion may be of limited use.
- Natural host range and symptom phenotype: when this information is available it may be used to differentiate between species, but its most common use will be to distinguish between strains belonging to the same species.