Family: Geminiviridae
Genus: Capulavirus
Distinguishing features
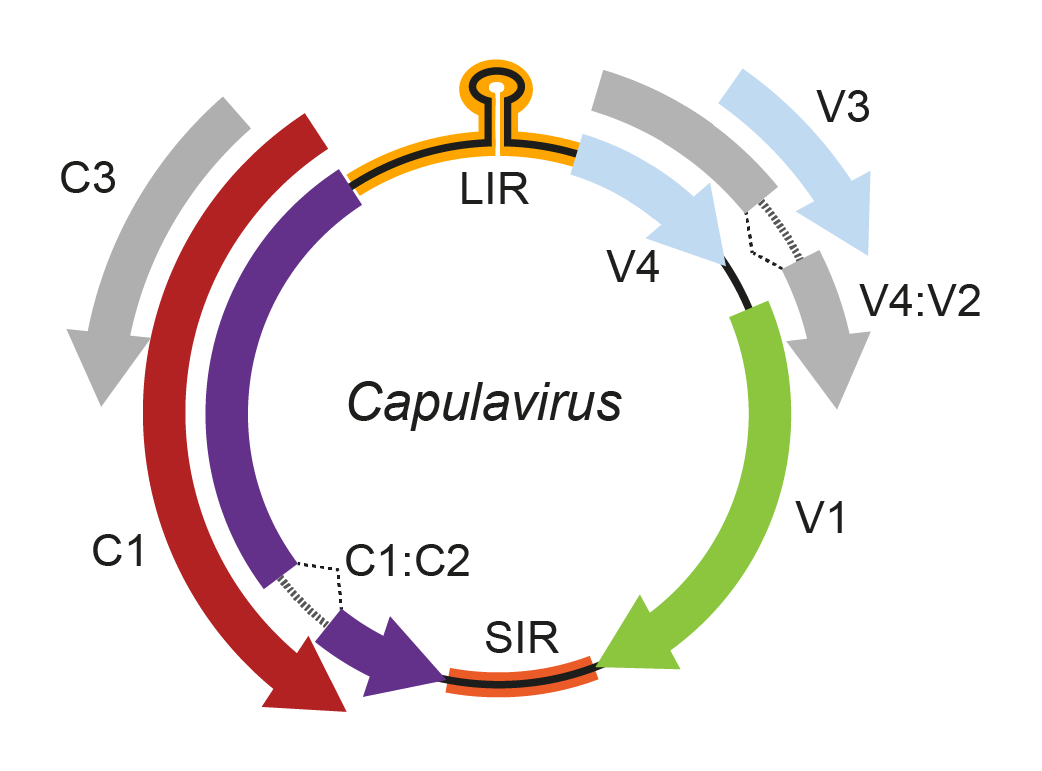
Uniquely among geminiviruses, capulavirus genomes have a complex arrangement of possible movement protein (MP)-encoding ORFs located in a 5′-direction from the coat protein (CP) ORF (Figure 1. Capulavirus). Two or more of these ORFs may constitute an intron-containing MP (Figure 1. Capulavirus). Geminate particles have been observed in purified preparations of Euphorbia caput-medusae latent virus by transmission electron microscopy (Roumagnac et al., 2015). Three capulaviruses have been shown to be transmitted by aphids (Roumagnac et al., 2015, Susi et al., 2019, Ryckebusch et al., 2020). All known capulaviruses have the nonanucleotide motif 5′-TAATATTAC-3′ at their presumed origins of virion strand replication.
|
|
|
Figure 1. Capulavirus. Genomic organization of capulaviruses. ORFs are denoted as being encoded on the virion-sense (V) or complementary-sense (C) strand. The position of the stem-loop containing the conserved 5′-TAATATTAC-3′ sequence located in the long intergenic region (LIR) is shown. Introns are predicted to occur between ORFs C1 and C2 and between ORFs V2 and V4. SIR, short intergenic region. |
Virion
See discussion under family description.
Genome organization and replication
Two overlapping complementary sense ORFs (C1 and C2) are predicted to encode the replication-associated protein (Rep) from a spliced transcript. As predicted for mastreviruses, it is possible that capulaviruses express a RepA protein from an unspliced transcript. The deduced amino acid sequences of capulavirus Rep proteins contain canonical rolling circle replication motifs, the ATPase motifs Walker-A, -B, and -C (related to Rep helicase activity) and the "geminivirus Rep sequence" (GRS) domain identified in other geminivirus Reps. However, they lack the canonical LXCXE retinoblastoma binding domain (Bernardo et al., 2013). A large complementary-sense ORF (C3) is completely embedded within the C1 ORF.
Capulaviruses have four virion sense ORFs: V1, V2, V3 and V4. Based on similarity with other geminiviruses, the V1 ORF is likely to encode the coat protein. The presence of transmembrane domains in the V3 and V4 ORFs, similar to those found in the MP of mastreviruses, suggest that these two ORFs may also encode MPs (Bernardo et al., 2013). The V2 ORF may not be functional, or may be expressed from a spliced transcript that would eliminate its start codon and introduce a frameshift that would cause the V2 and V4 ORFs to be expressed as a single protein (Bernardo et al., 2013, Bernardo et al., 2016).
Biology
Host range
Besides its natural host, Euphorbia caput-medusae, the experimental host range of Euphorbia caput-medusae latent virus includes Nicotiana benthamiana and Solanum lycopersicum (tomato) (Bernardo et al., 2013). Alfalfa leaf curl virus naturally infects Medicago sativa (alfalfa) (Bernardo et al., 2016), and can also experimentally infect Vicia faba (faba bean) and Nicotiana benthamiana (Roumagnac et al., 2015). French bean severe leaf curl virus and Plantago lanceolata latent virus are known to infect only Phaseolus vulgaris (bean) and Plantago lanceolata, respectively.
Transmission
Three members of the genus Capulavirus have been shown to be transmitted by aphids. Members of Alfalfa leaf curl virus and Euphorbia caput-medusae latent virus are transmitted by Aphis craccivora (Roumagnac et al., 2015, Ryckebusch et al., 2020) and members of Plantago lanceolata latent virus by Dysaphis plantaginea (Susi et al., 2019). No vector has been identified for viruses of the other species in the genus.
Species demarcation criteria
Nucleotide sequence identity: based on the distribution of pairwise identities of known capulavirus genomes, a species demarcation threshold of 78% is currently adopted (Varsani et al., 2017). Thus, any two capulaviruses whose genome sequences have a pairwise identity value above 78% should be considered isolates of the same species.