Family: Geminiviridae
Elvira Fiallo-Olivé, Jean-Michel Lett, Darren P. Martin, Philippe Roumagnac, Arvind Varsani, F. Murilo Zerbini and Jesús Navas-Castillo
The citation for this ICTV Report chapter is the summary published as Fiallo-Olivé et al.,
ICTV Virus Taxonomy Profile: Geminiviridae 2021, Journal of General Virology, 2021;102:001696
Corresponding authors: Elvira Fiallo-Olivé ([email protected]) and Jesús Navas-Castillo ([email protected])
Edited by: Hélène Sanfaçon, Andrew J. Davison and Luisa Rubino
Posted: March 2017, updated September 2018, August 2020 and October 2021
PDF: ICTV_Geminiviridae.pdf (August 2020 version)
Summary
The geminiviruses are a family of small, non-enveloped viruses with genomes comprising one or two single-stranded, circular DNAs of 2.5–5.2 kb. Geminiviruses infect a wide range of plant species and are transmitted by various insects in four families of homopterans (whiteflies, leafhoppers, aphids and treehoppers). Geminiviruses are important plant pathogens causing economically important diseases in most tropical and subtropical regions of the world.
Table 1. Geminiviridae. Characteristics of members of the family Geminiviridae.
| Characteristic | Description |
|---|---|
| Example: | bean golden yellow mosaic virus-[Dominican Republic-1987] (DNA-A: L01635, DNA-B: L01636), species Begomovirus birdi |
| Virion | Twinned (geminate) incomplete icosahedra, T=1, 22×38 nm with a single coat protein |
| Genome | 2.5–5.2 kb of single-stranded, circular DNA, mono- or bipartite |
| Replication | Complementary strand synthesized in the nucleus by host replication factors; double-stranded circular molecules serve as templates for both transcription and replication; replication employs a rolling-circle mechanism and also a recombination-dependent mechanism |
| Translation | From transcribed mRNAs; members of some genera use transcript splicing |
| Host range | Plants (monocots and dicots) |
| Taxonomy | Realm Monodnaviria, kingdom Shotokuvirae, phylum Cressdnaviricota, class Repensiviricetes, order Geplafuvirales: 15 genera collectively containing 549 species |
Viruses classified into the 15 genera show distinct host ranges, insect vectors and genome organizations:
Becurtovirus. This genus includes three species, Beet curly top Iran virus, Exomis microphylla latent virus (Claverie et al., 2018) and Spinach curly top Arizona virus (Varsani et al., 2014b). Becurtoviruses are monopartite viruses infecting dicot plants and members of at least two species are transmitted by leafhoppers. Members are unusual in that instead of the 5′-TAATATTAC-3′ nonanucleotide found at the origin of virion strand replication (v-ori) in almost all other geminiviruses, they have a 5′-TAAGATTCC-3′ nonanucleotide.
Begomovirus. This genus includes 445 species, members of which infect dicot plants and are transmitted by whiteflies of the Bemisia tabaci cryptic species complex (Brown et al., 2015). The genome can be either bipartite (two circular ssDNA molecules, each about 2.6 kb) or monopartite (a single approximately 2.7 kb circular ssDNA molecule) (Rojas et al., 2005). Begomoviruses usually induce severe symptoms in their hosts, including yellow mosaic, golden mosaic and leaf curl. Devastating pathogens include members of the species African cassava mosaic virus, Bean golden mosaic virus, Cotton leaf curl Multan virus and Tomato yellow leaf curl virus. It has been demonstrated for a begomovirus, tomato yellow leaf curl virus, that in addition to the host plants its replication also occurs in the salivary glands of the insect vector B. tabaci (He et al., 2020). Three classes of circular DNA satellites have been described associated with begomoviruses: betasatellites, alphasatellites and deltasatellites (Zhou 2013, Lozano et al., 2016).
Capulavirus. This genus includes four species, Alfalfa leaf curl virus (Roumagnac et al., 2015), Euphorbia caput-medusae latent virus (Bernardo et al., 2013), French bean severe leaf curl virus and Plantago lanceolata latent virus (Susi et al., 2017). Members of three species are transmitted by aphids (Roumagnac et al., 2015, Susi et al., 2019, Ryckebusch et al., 2020). A unique feature of capulavirus genomes is the complex arrangement of possible movement protein-encoding ORFs located in a 5′-direction from the coat protein gene. All known capulaviruses have a monopartite genome and the 5′-TAATATTAC-3′ nonanucleotide at the v-ori.
Citlodavirus. This genus includes four species, Camellia chlorotic dwarf-associated virus (Zhang et al., 2018), Citrus chlorotic dwarf associated virus (Loconsole et al., 2012), Paper mulberry leaf curl virus 2 (Qiu et al., 2020) and Passion fruit chlorotic mottle virus (Fontenele et al., 2018). Members of these species have been isolated from symptomatic dicot host plants, both trees and shrubs. All citlodaviruses are monopartite and their genomes contain the 5′-TAATATTAC-3′ nonanucleotide at the v-ori.
Curtovirus. This genus includes three recognised species, Beet curly top virus (Stanley et al., 1986), Horseradish curly top virus (Klute et al., 1996) and Spinach severe curly top virus (Hernandez and Brown 2010). Members infect dicot plants and are transmitted by leafhoppers. In this genus, the most studied virus is beet curly top virus an economically important pathogen in North America and Iran with a wide host range (Chen and Gilbertson 2009)
Eragrovirus. This genus includes the single species, Eragrostis curvula streak virus, members of which have monopartite genomes (Varsani et al., 2014b). All known isolates have been found infecting the monocot plant Eragrostis curvula (weeping love grass) in the Kwa-Zulu Natal region of South Africa (Varsani et al., 2009). Like the becurtoviruses, eragroviruses have a 5′-TAAGATTCC-3′ nonanucleotide sequence at the v-ori.
Grablovirus. This genus includes three species, Grapevine red blotch virus (Krenz et al., 2012), Prunus latent virus (Al Rwahnih et al., 2018) and Wild Vitis latent virus (Perry et al., 2018). Grapevine red blotch virus is transmitted by a treehopper (Bahder et al., 2016). Members have a 5′-TAATATTAC-3′ nonanucleotide at the v-ori and infect dicot plants.
Maldovirus. This genus includes three species, Apple geminivirus 1 (Liang et al., 2015), Grapevine geminivirus A (Al Rwahnih et al., 2017) and Juncus maritimus geminivirus 1 (Claverie et al., 2018). Members of the mentioned species have been described from asymptomatic plants and infect dicot (apple, grapevine) and monocot (Juncus maritimus) plants. All maldovirus genomes contain the origin of replication (5′-TAATATTAC-3′) conserved in most geminiviruses.
Mastrevirus. Mastreviruses have monopartite genomes, infect mostly monocots and some dicots, and are transmitted by different species of leafhoppers (Muhire et al., 2013). Of the 45 species included in this genus, the best studied are Maize streak virus, members of which cause a severe disease of maize in Africa (Shepherd et al., 2010), and Wheat dwarf virus, members of which cause a disease of wheat across northern Europe, the Middle East and Asia (Kvarnheden et al., 2002). A member of the species Wheat dwarf India virus has been shown to associate with alphasatellites and betasatellites (Kumar et al., 2014).
Mulcrilevirus. This genus includes two species, Mulberry crinkle leaf virus (Lu et al., 2015, Ma et al., 2015) and Paper mulberry leaf curl virus 1 (Qiu et al., 2020). Members of the genus contain monopartite genomes with origin of replication (5′-TAATATTAC-3′) conserved in most geminiviruses. Mulcrileviruses have been described from symptomatic plants (Morus alba or Broussonetia papyrifera) from China. An isolate of mulberry crinkle leaf virus is transmitted by the leafhopper Tautoneura mori (Lu et al., 2021).
Opunvirus. The single member of this genus is Opuntia virus 1. Isolates of Opuntia virus 1 have been described from asymptomatic New World Cactaceae plants (belonging to 20 different cactus species from the subfamilies Cactoideae and Opuntioideae) and from cactus-feeding cochineal insects (Dactylopius sp.) (Fontenele et al., 2020). Opuntia virus 1 genomes are monopartite and contain the conserved geminivirus nonanucleotide (5′-TAATATTAC-3′).
Topilevirus. This genus includes two species, Tomato apical leaf curl virus (Vaghi Medina et al., 2018) and Tomato geminivirus 1 (Fontenele et al., 2017). Members of these species contain monopartite genomes with the conserved geminivirus nonanucleotide (5′-TAATATTAC-3′). Both viruses infect the same host plant, tomato, and tomato geminivirus 1 has also been isolated from Cleome sp. plants.
Topocuvirus. Members of the single species in this genus, Tomato pseudo-curly top virus, are transmitted by a treehopper (Briddon et al., 1996). The virus genome is similar to those of curtoviruses.
Turncurtovirus. There are currently three species in this genus, Turnip curly top virus (Varsani et al., 2014b, Razavinejad et al., 2013), Turnip leaf roll virus (Kamali et al., 2016) and Sesame curly top virus (Hasanvand et al., 2018). Members are monopartite. Isolates of turnip curly top virus and turnip leaf roll virus have only been found so far in Iran and are transmitted by the leafhopper Circulifer haematoceps. Isolates of sesame curly top virus have been identified from sesame (Sesamum indicum) in Pakistan and Iran and from the leafhopper Circulifer haematoceps collected from sesame plants also in Iran.
Virion
Morphology
Virions are typically twinned ("geminate"). For maize streak virus (MSV, genus Mastrevirus), cryo-electron microscopy has shown that virions are about 22×38 nm in size, consisting of two incomplete icosahedra (T=1) containing a total of 110 coat protein subunits organized as 22 pentameric capsomers (Zhang et al., 2001). The structure of Ageratum yellow vein virus (genus Begomovirus) at 3.3 Å resolution has shown that the N-terminus of the single CP adopts three different conformations essential for building the interface between geminate halves (Hesketh et al., 2018) (Figure 1. Geminiviridae).
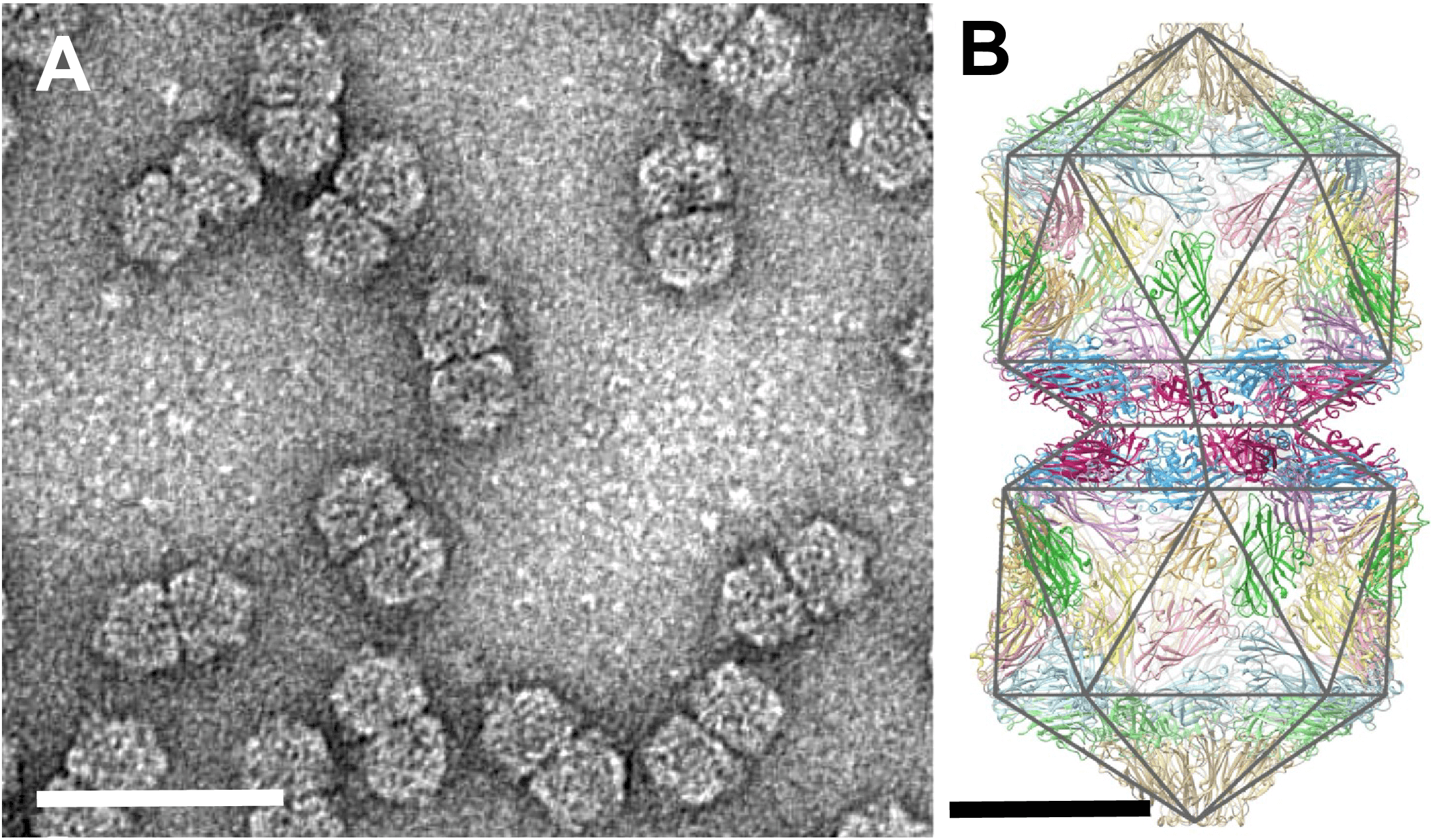 |
| Figure 1. Geminiviridae. (A) Purified particles of maize streak virus (MSV) stained with uranyl acetate showing typical twinned quasi-isometric subunits. The bar represents 50 nm. Reproduced with permission from (Zhang et al., 2001). (B) Complete atomic model for all 110 subunits in the capsid of Ageratum yellow vein virus (AYVV), with a polyhedral cage showing the symmetry of the particle. The fivefold symmetry axis is vertical and through the center of the particle in this view. Bar, 10 nm. CC BY 4.0 licence from (Hesketh et al., 2018). |
Physicochemical and physical properties
The virion S20,w is approximately 70S (Goodman et al., 1980).
Nucleic acid
Twinned virions contain a single copy of circular ssDNA, ranging from 2.5 to 3.2 kb. Hence, for viruses with bipartite genomes, two virions containing different genomic components are required for infection, the total genome being about 5.2 kb (Von Arnim et al., 1993, Hamilton et al., 1983). Half-size defective components and ssDNA satellites are also encapsidated (Zhou 2013).
Proteins
Virions contain a single structural protein (CP; Mr about 28–34×103). No other proteins have been found associated with virions (Zhang et al., 2001, Goodman et al., 1980).
Genome organization and replication
Viruses in the genera Becurtovirus, Capulavirus, Citlodavirus, Curtovirus, Eragrovirus, Grablovirus, Maldovirus, Mastrevirus, Mulcrilevirus, Opunvirus, Topilevirus, Topocuvirus and Turncurtovirus have a single genomic component, whereas those in the genus Begomovirus have either one or two components (Figure 2. Geminiviridae).
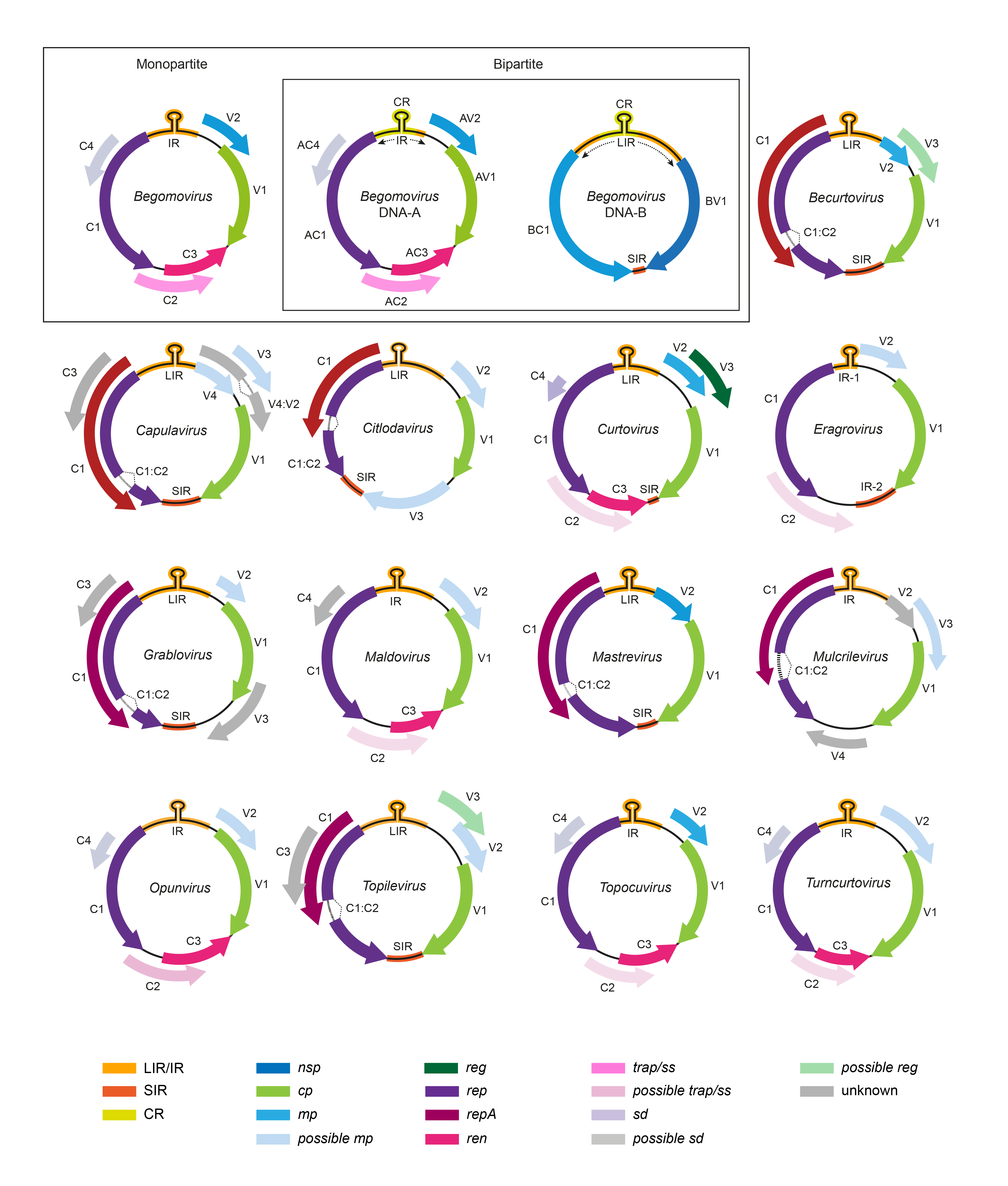 |
| Figure 2. Geminiviridae. Genomic organization of members of the family Geminiviridae. ORFs are denoted as being encoded on the virion-sense (V) or complementary-sense (C) strand, and corresponding protein products are coded by colour. The position of the stem-loop containing the conserved 5′-TAATATTAC-3′ sequence located in the intergenic region (IR/IR-1) and long intergenic region (LIR) is shown. For begomoviruses, V2/AV2 is not present in New World viruses. CP, coat protein; IR, intergenic region; LIR, long intergenic region; SIR, short intergenic region; MP, movement protein; NSP, nuclear shuttle protein; REn, replication enhancer protein; Rep, replication-associated protein; SIR, short intergenic region; TrAP, transcriptional activator protein. |
In the host plants, replication occurs through double-stranded replicative intermediates by both recombination-dependent and rolling circle mechanisms (Stenger et al., 1991, Preiss and Jeske 2003). Complementary-sense DNA synthesis on the virion-sense (encapsidated) strand to produce dsDNA depends solely on host factors (Saunders et al., 1992). Virus ssDNA synthesis is initiated by cleavage of the virion-sense strand by the virus-encoded replication-associated protein (Rep) immediately downstream of the 3′-thymidine residue in a conserved 5′-TARTATT↓AC-3′ sequence located in the loop of a potential stem-loop structure within the intergenic region (IR) (Laufs et al., 1995, Fontes et al., 1994). Large amounts of heterogenous sub- and extra-genome length geminivirus DNA (hDNA) are generated during rolling circle replication, and most of the viral DNA that accumulates within infected cells is likely produced from this hDNA by recombination-dependent mechanisms that rely on host DNA double-stranded break repair complexes (Preiss and Jeske 2003). Geminiviruses do not encode a DNA polymerase, and consequently rely entirely on host factors that must be recruited during the early stages of replication. One member of the genus Begomovirus, tomato yellow leaf curl virus, is replicated in the insect vector Bemisia tabaci. The viral Rep interacts with whitefly PCNA, which recruits DNA Polδ for virus replication (He et al., 2020).
In all cases, coding regions in both virion-sense and complementary-sense strands diverge from an intergenic region, and transcription is bi-directional, with independently controlled transcripts initiating within the intergenic region (IR) (Sunter and Bisaro 1989). Geminiviruses use multiple overlapping transcripts to control gene expression; those in the genera Mastrevirus (Schalk et al., 1989), Citlodavirus and Mulcrilevirus (Qiu et al., 2020) and, probably, also in the genera Becurtovirus, Capulavirus, Grablovirus and Topilevirus, additionally use transcript splicing.
Biology
Members of the genus Begomovirus are transmitted by whiteflies, those of the genera Mastrevirus, Curtovirus, Becurtovirus, Mulcrilevirus and Turncurtovirus are transmitted by leafhoppers, those of the genus Capulavirus are transmitted by aphids, and members of species in the genera Topocuvirus and Grablovirus are transmitted by treehoppers. The vectors of members of the other genera remain unknown. Some viruses in the family Geminiviridae infect a wide range of plant species while others have a narrow host range. Members of the genera Becurtovirus, Begomovirus, Capulavirus, Citlodavirus, Curtovirus, Grablovirus, Mulcrilevirus, Opunvirus, Topilevirus, Topocuvirus and Turncurtovirus infect dicot plants. However, most members of the genus Mastrevirus, the only member of the genus Eragroviurs and one of the three members of the genus Maldovirus infect monocot plants. Geminiviruses are important plant pathogens causing economically important diseases in most tropical and subtropical regions of the world. Further information is provided on individual genus pages.
Derivation of names
Becurtovirus: from the species Beet curly top Iran virus.
Begomovirus: from the species Bean golden yellow mosaic virus (previously Bean golden mosaic virus).
Capulavirus: from the species Euphorbia caput-medusae latent virus.
Citlodavirus: from the species Citrus chlorotic dwarf associated virus.
Curtovirus: from the species Beet curly top virus.
Eragrovirus: from the species Eragrostis curvula streak virus.
Geminiviridae: from Latin geminus meaning "twin", describing the characteristic twinned (geminate) particle morphology.
Grablovirus: from the species Grapevine red blotch virus.
Maldovirus: from the scientific name of the host plant Malus domestica, and virus.
Mastrevirus: from the species Maize streak virus.
Mulcrilevirus: from the species Mulberry crinkle leaf virus.
Opunvirus: from the species Opuntia virus 1.
Topilevirus: from the species Tomato apical leaf curl virus.
Topocuvirus: from the species Tomato pseudo-curly top virus.
Turncurtovirus: from the species Turnip curly top virus.
Genus demarcation criteria
These include host range (monocots or dicots), type of vector (leafhoppers, treehoppers, whiteflies, aphids), genome organization (mono- or bipartite), and phylogenetic relationships.
Relationships within the family
Phylogenetic analysis of complete genome sequences (DNA-A sequences in the case of bipartite begomoviruses) from isolates of representative species shows that geminiviruses cluster into groups corresponding to the 14 genera (Figure 3. Geminiviridae). In addition, they cluster according to geographic distribution, at least within the begomoviruses, probably reflecting their evolutionary divergence as a consequence of isolation due to the inability of their insect vectors to fly long distances (Rocha et al., 2013). Despite frequent inter-species recombination events and the increasing worldwide movement of infected plants, it is remarkable that this geographical distribution is still apparent.
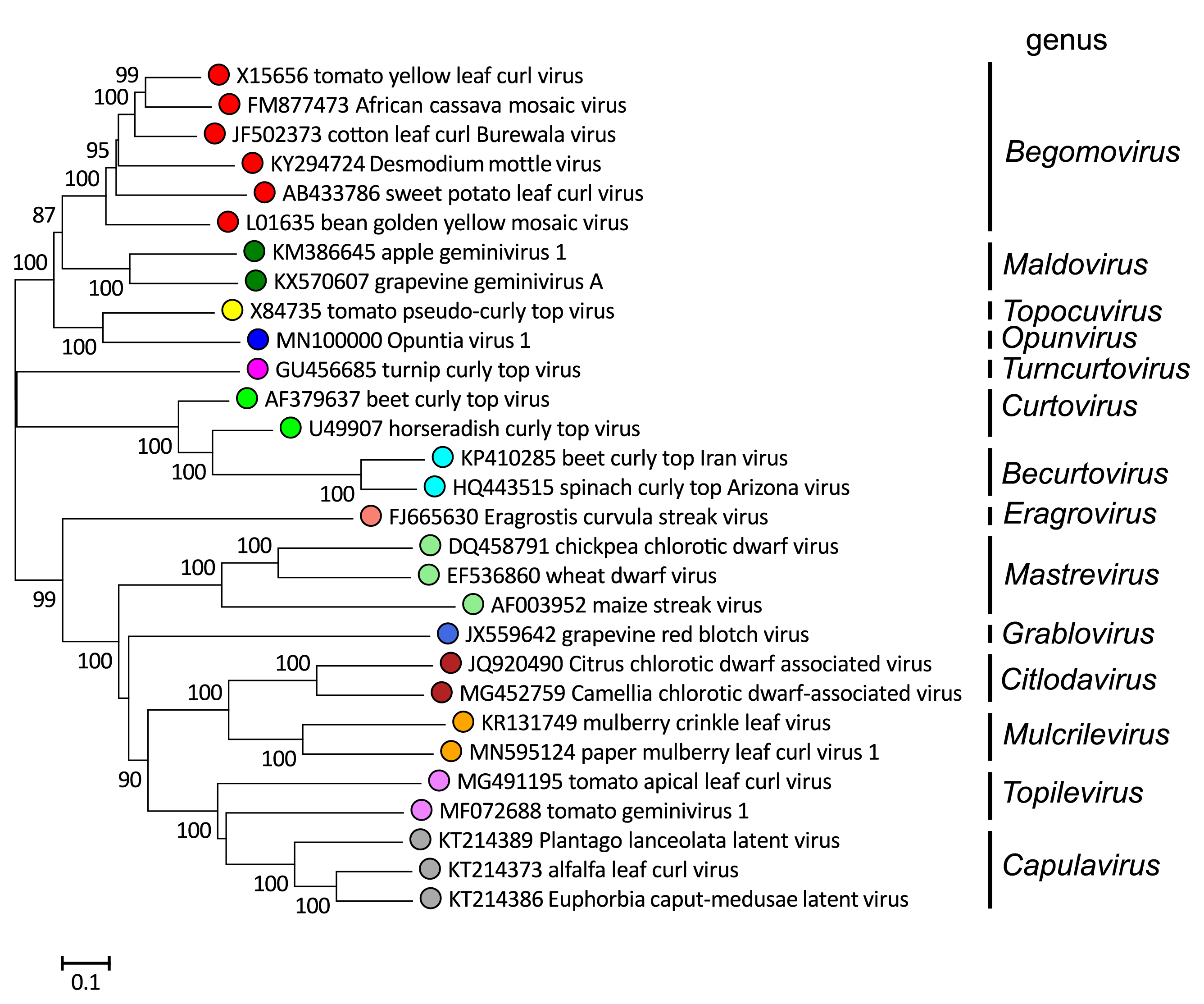 |
| Figure 3. Geminiviridae. Unrooted, neighbor-joining phylogenetic tree of the full genome sequences (or DNA-A sequences for bipartite geminiviruses) of representative isolates in the genera in the family Geminiviridae. This tree is presented as a guide to describe relative similarities between the geminivirus groups, as it is not possible to align all of the representative genomes from the fourteen genera, as inter-species and inter-genus recombination has played a major part in the diversification of the represented virus isolates. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the plant virus families Geminiviridae and Nanoviridae have circular ssDNA genomes and replicate by a rolling circle mechanism. All these viruses have highly conserved sequences (5′-TARTATTAC-3′ (geminiviruses), 5′-TAGTATTAC-3′ (nanovirids)) in the loop of a putative stem-loop structure within the IR, in which a nick is introduced during the initiation of replication. Interestingly, the majority of alphasatellites that associate with begomoviruses have the nanovirus-like sequence 5′-TAGTATTAC-3′, consistent with them having evolved from components of nanoviruses. Similar structures are found in members of the animal virus families Circoviridae and Anelloviridae, and the fungus virus family Genomoviridae, all of them also havingsmall circular ssDNA genomes. It is speculated that geminiviruses derive from prokaryotic episomal replicons, based on conservation of motifs in proteins that function in rolling circle replication initiation.

