Family: Caulimoviridae
Genus: Tungrovirus
Distinguishing features
Rice tungro bacilliform virus (RTBV) is a member of the only species in the genus Tungrovirus. Along with members of the genus Badnavirus, RTBV is readily distinguished from other members of the Caulimoviridae by its bacilliform-shaped virions. Although the genome organization of RTBV is very similar to that of members of the genus Badnavirus, it differs by the presence of a fourth ORF (ORF4). This fourth ORF is expressed from a monocistronic RNA that is produced by splicing of the pregenomic RNA. RTBV can also be differentiated from members of the genus Badnavirus by phylogenetic placement based on analysis of polymerase gene sequences.
Virion
Morphology
Virions are bacilliform with a width of 30 nm and a predominant length of 130 nm; longer virions, up to 300 nm, are found in some isolates. The virion structure is based on a T=3 icosahedron cut across its three-fold axis, the tubular portion being made up of rings of hexamer subunits with a repeat distance of about 10 nm.
Physicochemical and physical properties
Virions sediment with an S20,w of about 200 S and have a buoyant density in CsCl of about 1.36 g cm−3.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of about 8.0 kbp. Each strand of the genome has a single discontinuity.
Proteins
Protein precursors in the ORF3 polyprotein are processed through the action of the aspartic protease (AP). The mature capsid protein (CP) has been mapped to aa 477–791 (GenBank accession X57924), giving rise to a protein of 37.3 kDa. The aspartic protease has been mapped to aa 965–1085, giving rise to a protein of 13.8 kDa.
Genome organization and replication
The genome contains four ORFs, a large intergenic region between ORF4 and ORF1 and a small intergenic region between ORF3 and ORF4 (Figure 1 Tungrovirus). The pregenomic RNA promoter, polyadenylation signal and negative-sense strand primer-binding site are all located in the large intergenic region. The function of protein P1 is unknown, P2 is the virion-associated protein, and P3 is a polyprotein with movement protein, coat protein, aspartic protease and RT/RNase H1 domains, in that order. P4 is a suppressor of cell-to-cell spread of silencing (Rajeswaran et al., 2014).
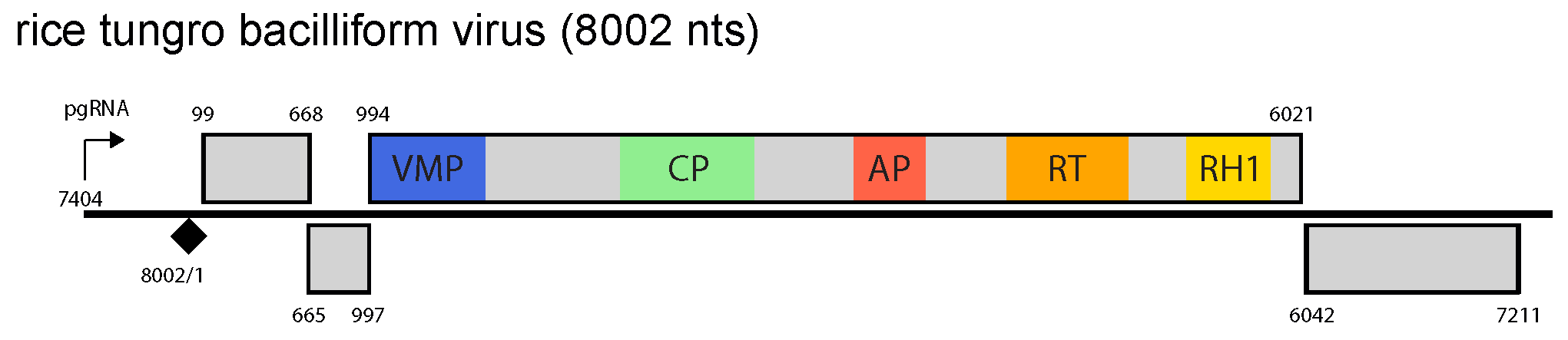 |
| Figure 1 Tungrovirus. Tungrovirus genome organization. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999)and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (pepsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. |
A single, greater-than-genome length, terminally redundant pregenomic RNA is transcribed. The pregenomic RNA serves as a polycistronic mRNA for translation of ORFs 1–3. Translation of ORF1 is initiated by ribosome shunting and translation of ORF2 and ORF3 by leaky scanning. Consistent with a leaky scanning model of translation, the start codon of ORF1 is non-conventional (AUU), the start codon of ORF2 is in an unfavorable translation context and there is an absence of internal AUG codons in both ORFs. Furthermore, ORF3 is in a −1 translational frame relative to ORF2, which in turn is in a −1 translational frame relative to ORF1. The termination codon of ORF1 overlaps with the start codon of ORF2 (ATGA), and similarly for the termination codon of ORF2 and the start codon of ORF3.
ORF4 is expressed from a monocistronic RNA that is produced by splicing of the pregenomic RNA. An intron of 6,300 nt is excised in a way that fuses the first small ORF (sORFA) in the leader sequence of the pregenomic RNA in frame to ORF4 at a position that is 22 codons upstream of the start codon of this ORF.
Biology
RTBV is transmitted by leafhoppers in the genera Nephotettix and Recilia. Although able to replicate independently in its host, RTBV is only able to be transmitted when the leafhopper has acquired rice tungro spherical virus (RTSV; family Secoviridae, genus Waikavirus) simultaneously or previously. This suggests that RTSV may contribute a helper component required for RTBV transmission. RTBV is not mechanically transmissible. There is no evidence of seed transmission. Host range is limited to members of the Poaceae and Cyperaceae.
Antigenicity
RTBV is moderately immunogenic.
Derivation of names
Tungrovirus: derived from tungro, meaning “degenerated growth” in a Filipino dialect.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Host ranges
- Differences in polymerase (RT + RNAse H) nt sequences of more than 20%

