Family: Caulimoviridae
Genus: Badnavirus
Distinguishing features
The genera Badnavirus and Tungrovirus differ from other genera in the family Caulimoviridae in that their members have bacilliform-shaped virions. Members of the genus Badnavirus can be distinguished from members of the species Tungrovirus oryzae, the sole member of the genus Tungrovirus, by genome organization, the lack of any RNA splicing during replication, the lack of dependency on a helper virus for vector transmission, and phylogenetic placement in reverse transcriptase domain-based trees.
Virion
Morphology
Virions are bacilliform with parallel sides and rounded ends (Figure 1.Caulimoviridae). Virions are uniformly 30 nm in width. The modal particle length is 130 nm, but virions ranging in length from 60 to 900 nm are commonly observed. No projections, or other capsid surface features, have been observed by electron microscopy. The tubular portion of the virion has a structure based on an icosahedron cut across its 3-fold axis, with a structural repeat of 10 nm and nine rings of hexamer subunits per 130 nm length.
Physicochemical and physical properties
Purified virions have an A260/280nm ratio of 1.26 (uncorrected for light scattering).
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of 7.2–9.2 kbp. Each strand of the genome has a single discontinuity.
Proteins
Virions have two proteins, the capsid protein (CP) and the virion-associated protein (VAP). Three domains are recognized in the CP: the N-terminal intrinsically disordered (NID) domain, the capsid (CA) domain and the nucleocapsid (NC) domain (Vo et al., 2016). The NC domain contains a zinc finger knuckle at the C-terminus and a basic, intrinsically disordered region at the N-terminus. Two isoforms of the CP (CP1 and CP2) are observed, which are thought to differ in length of the NID domain either because of the presence of alternative aspartic protease (AP) cleavage sites or as a result of targeted degradation of the larger CP isoform (CP1). The immunodominant linear epitopes are located in the NID domain.
Proteins within the P3 polyprotein are cleaved through the action of the AP but enzyme substrate sites, and hence boundaries of the mature proteins, are unknown except for the N-terminus of the smaller CP isoform (CP2) of banana streak MY virus (BSMYV) and banana streak OL virus (BSOLV). The N-terminus of the CP1 isoform has not been sequenced because of chemical blocking.
The movement protein (MP) belongs to the 30K superfamily of viral movement proteins, which form tubular structures that occupy the plasmodesmata and increase the size exclusion limit for passage of macromolecules. Electron microscopic studies suggest that whole virions traffic between the cells utilizing the tubules (Cheng et al., 1998).
Genome organization and replication
The genome contains three ORFs coding for proteins denominated P1–P3 (Figure 1 Badnavirus), except for sweet potato pakakuy virus and Jujube mosaic-associated virus, where ORF3 is divided into two, with a short intergenic region. The function of protein P1 is unknown, P2 is the virion-associated protein, P3 is a polyprotein with movement protein, capsid protein, aspartic protease and RT/RNase H1 protein precursors, in that order.
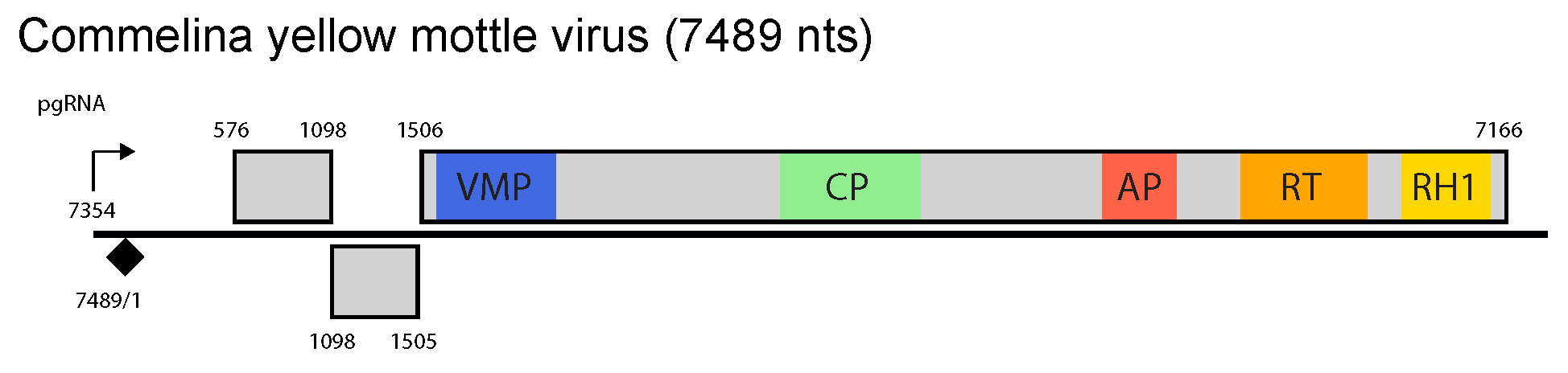
Figure 1 Badnavirus. Badnavirus genome organization. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999) and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (p epsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. Note that in Commelina yellow mottle virus, an authentic start codon of ORF1 is the second in-frame AUG at position 576 (Cheng et al., 1996), consistent with position of the ribosome shunt landing site (Pooggin et al., 1999).
A single, greater-than-genome length, terminally redundant pregenomic RNA is transcribed. No subgenomic RNAs have been observed. The pregenomic RNA serves as a polycistronic mRNA for translation of the three ORFs. By analogy to rice tungro bacilliform virus (RTBV), translation of ORF1 is initiated by ribosome shunting and translation of ORF2 and ORF3 by leaky scanning. Consistent with a leaky scanning model of translation, the start codon of ORF1 and ORF2 are in unfavourable translation contexts and there is paucity of internal AUG codons in both ORFs. In BSMYV and banana streak VN virus (BSVNV), ORF1 begins with a nonconventional start codon (CUG) (Geering et al., 2005, Lheureux et al., 2007). Furthermore, ORF3 is in a −1 translational frame relative to ORF2, which in turn is in a −1 translational frame relative to ORF1. The termination codon of ORF1 overlaps with the start codon of ORF2 (ATGA), and similarly for the termination codon of ORF2 and the start codon of ORF3.
Sites where the ORF3 polyprotein is cleaved by the viral aspartic protease have not been determined.
Biology
Transmission is in a semi-persistent manner by mealybugs and, for some members, by aphids or lace bugs. The virus does not multiply in its mealybug vector and there is no transovarial transmission. All motile life stages of vectors can acquire and transmit the virus. There is little information on the possible transmission of badnaviruses by other vector types. Seed transmission at a rate of 30–63% has been recorded for Kalanchoe top-spotting virus (KTSV). KTSV, cacao swollen shoot virus and an unidentified badnavirus from sugarcane have been mechanically transmitted but attempts with other viruses have been unsuccessful, which may relate to the presence of inhibitory substances in the plant sap or low virus titers. Some badnaviruses infecting woody hosts have been transmitted by dodder and grafting. Biological host ranges are narrow and restricted to one or two plant families.
Replication-competent endogenous badnaviral forms belonging to the species Banana streak OL virus, Banana streak GF virus and Banana streak IM virus are present in the genome of the wild type banana species Musa balbisiana (Chabannes et al., 2013). These endogenous elements are responsible for causing infection in M. acuminata × M. balbisiana hybrids, especially following plant propagation by tissue culture. Many other plant species contain endogenous badnaviral elements, but most are probably replication-defective (Teycheney and Geering 2011).
Antigenicity
Serological relationships between members of different species are highly variable ranging from moderate to strong.
Derivation of names
Badnadvirus: derived from bacilliform DNA virus.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Host ranges
- Differences in polymerase (RT + RNAse H) nt sequences of more than 20%
- Vector specificities
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Aucuba bacilliform virus | AuBV | |
| banana streak CA virus | HQ593111 | BSCAV |
| banana streak UJ virus | BSUJV | |
| banana streak UK virus | BSUKV | |
| banana streak PE virus | MN187554 | BSPEV |
| enset leaf streak virus | MF991909 | ELSV |
| Epiphyllum badnavirus 1 | MH396440 | EpBV1 |
| Hibiscus bacilliform virus | KF875586 (NC_023485) | HiBV |
| Mimosa bacilliform virus | MBV | |
| Pelargonium vein banding virus | GQ428155 (NC_13262) | PVBV |
| Stilbocarpa mosaic bacilliform virus | AF478961* | SMBV |
| sweet potato badnavirus B | FJ560944 (NC_012728) | |
| Yucca bacilliform virus | AF468688 | YBV |
Virus names and virus abbreviations are not official ICTV designations.
* Sequence does not comprise the complete genome.
† Sequence amplified by PCR from total plant DNA extract; no experimental proof that the sequence derives from an exogenous viral genome.

