Family: Caulimoviridae
Genus: Dioscovirus
Distinguishing features
Dioscorea nummularia-associated virus (DNUaV) is a member of the only species in the genus. Its genome encodes four ORFs with ORF3 and ORF4 separated by an intergenic region, and ORF4 encoding a translation transactivator domain.
Virion
Virions are unknown.
Nucleic acid
The genome of Dioscorea nummularia-associated virus comprises 8139 bp with four ORFs.
Genome organization and replication
DNUaV has unique genome features. DNUaV encodes four ORFs with the size of ORFs 1 and 3 similar to that of both badnaviruses and tungroviruses, as are the arrangement of the movement protein, coat protein, aspartic protease and reverse transcriptase / ribonuclease H in ORF3 (Figure 1 Dioscovirus). The relative positions of ORF1 and ORF2 are similar to those of badnaviruses, while ORF2 and ORF3 overlap each other by 47 nt which is also similar to the badnaviruses cacao swollen shoot virus, gooseberry vein banding virus, Piper yellow mottle virus and sweet potato pakakuy virus. However, unlike those badnaviruses with a fourth ORF which always overlaps with ORF3, in DNUaV ORF4 is separated from ORF3 by a short intergenic region which is more similar to genome organization of RTBV, the sole member of the genus Tungrovirus. Further, the size of DNUaV ORF4 is also similar to that of rice tungro bacilliform virus (RTBV). However, unlike RTBV, the ORF4 gene product of DNUaV contains a conserved translation transactivator domain, which is typical of ORF6 of caulimoviruses and soymoviruses, and which is also present in ORF4 of cavemoviruses and solendoviruses. Unlike the DNUaV ORF4 sequence, the ORF4 sequences of both cavemoviruses and solendoviruses also include the conserved coiled-coil motifs characteristic of the virion-associated protein.
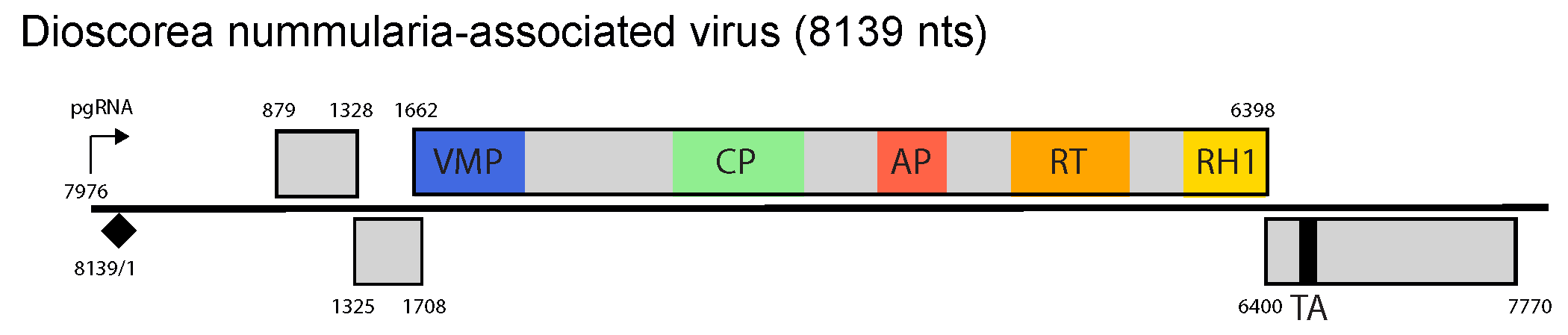 |
| Figure 1 Dioscovirus. Dioscovirus genome organization. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999) and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (pepsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. The conserved translation transactivator (TA) domain is shown in black. |
Biology
The only known host of DNUaV is Dioscorea nummularia.
Species demarcation criteria
DNUaV is the sole member of the genus.

