Family: Caulimoviridae
Genus: Cavemovirus
Distinguishing features
The members of this genus produce virions and cytoplasmic inclusions similar to those of viruses in the genera Caulimovirus, Petuvirus and Soymovirus but differ from them in genome organization and phylogenetic placement based on analysis of polymerase gene sequences. The order of the movement protein and coat protein is also reversed to that of all other genera in the Caulimoviridae.
Virion
Morphology
Virions are isometric and about 50 nm in diameter.
Physicochemical and physical properties
The sedimentation coefficient, S20,w, of cassava vein mosaic virus (CsVMV) is estimated to be 246 S.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of 7.7–8.2 kbp.
Genome organization and replication
Both cassava vein mosaic virus (CsVMV) and sweet potato collusive virus (SPCV) have four ORFs and a large intergenic region, in which the pregenomic RNA promoter, the RNA polyadenylation signal and the negative-sense strand primer-binding site are located (Figure 1.Cavemovirus). ORF1 of CsVMV and SPCV encode a polyprotein with coat protein and movement protein domains, in that order. The putative proteins encoded by CsVMV and SPCV ORF2 have no known function. For both viruses, the proteins encoded by ORF3 and ORF4 are the polymerase polyprotein [aspartic protease (AP), reverse transcriptase (RT) and RNase H1] and the virion-associated protein/translation transactivator, respectively.
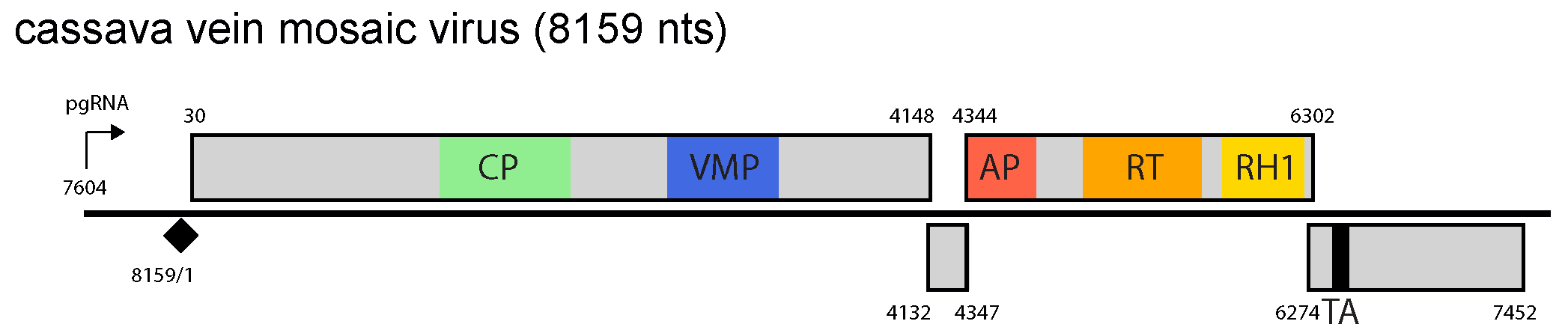 |
|
Figure 1. Cavemovirus. Cavemovirus genome organization. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999)and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (pepsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. The conserved translation transactivator (TA) domain is shown in black. |
The 5′-leader sequence of the pregenomic RNA folds into a stable hairpin structure preceded by a small ORF and thus a ribosome shunt mechanism of initiation of translation of ORF1 is assumed to be utilized (Pooggin et al., 1999). The presence of a translation transactivator homolog in the genome would suggest a similar mechanism of expression of downstream ORFs in a polycistronic mRNA as one utilized by members of the genus Caulimovirus.
Biology
The only known natural hosts of CsVMV and SPCV are cassava (Manihot esculenta) and sweet potato (Ipomoea batatas and I. setosa), respectively. There is no conclusive evidence of vector or seed transmission of CsVMV and SPCV. Furthermore, attempts at mechanical or aphid transmission of SPCV have been unsuccessful. Both viruses are transmitted by vegetative propagation.
Derivation of names
Cavemovirus: derived from cassava vein mosaic virus, member of the type species of the genus.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Host range
- Differences in polymerase (RT + RNAse H) nt sequences of more than 20%

