Family: Caulimoviridae
Genus: Solendovirus
Distinguishing features
Sweet potato vein clearing virus (SPVCV) and tobacco vein clearing virus (TVCV) are members of the only two species in the genus (Geering et al., 2010). They can be distinguished from other members of the Caulimoviridae by their phylogenetic placement using polymerase gene sequences.
Virion
Morphology
TVCV virions are isometric in shape and 48–50 nm in diameter.
Physicochemical and physical properties
TVCV virions have a buoyant density of 1.348 g cm−3 in CsCl.
Nucleic acid
TVCV and SPVCV virions contain a single molecule of non-covalently closed circular dsDNA of 7.7 kbp and 8.8 kbp, respectively.
Genome organization and replication
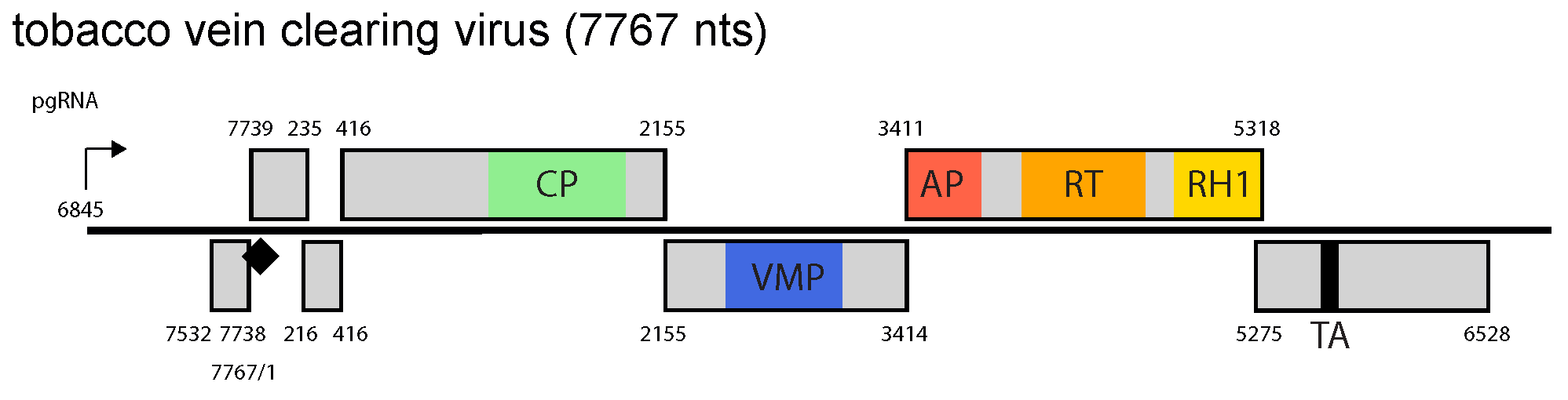
ORF1 codes for the capsid protein, ORF2 for a movement protein, ORF3 for a replicase polyprotein and ORF4 product is a transactivator (Figure 1.Solendovirus). Ribosome shunting and transactivator (TA)-mediated reinitiation of translation on the polycistronic pgRNA of TVCV are assumed to be utilized for gene expression (Pooggin and Ryabova 2018).
 |
|
Figure 1. Solendovirus. Solendovirus genome organization. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999)and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (pepsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. The conserved translation transactivator (TA) domain is shown in black. |
Biology
SPVCV is graft transmissible to the indicator plant Ipomea setosa. TVCV is not transmissible by aphid or mechanical inoculation; infection in the interspecific allohexaploid Nicotiana edwardsonii (N. clevelandii × N. glutinosa) is believed to originate from the activation of replication-competent endogenous TVCV (eTVCV) forms embedded in the nuclear genome of the plant (Lockhart et al., 2000). The virus is vertically transmitted to all progeny as part of the host’s genome. TVCV infections have only been observed in Nicotiana edwardsonii, although eTVCV sequences are present in Nicotiana sylvestris, Nicotiana tabacum, Nicotiana tomentosiformis, Solanum habrochaites and Solanum lycopersicum, and related sequences have been detected in several other plant species in the Solanaceae.
Derivation of names
Solendovirus: derived from the members of the family being endogenous viruses of plants in the family Solanaceae
Species demarcation criteria
The criteria demarcating species in the genus are:
- Host range
- Differences in polymerase (RT + RNAse H) nucleotide sequences of more than 20%

