Family: Caulimoviridae
Genus: Caulimovirus
Distinguishing features
Members of this genus have virions and cytoplasmic inclusions similar to those of members of the genera Cavemovirus, Petuvirus and Soymovirus, but differ from them in genome organization and phylogenetic placement based on analysis of polymerase gene sequences. Caulimovirus genomes have seven open reading frames and can be distinguished from those of their closest relatives, the soymoviruses, by the presence of only two ORFs between ORF1 (movement protein) and ORF4 (coat protein) instead of three. The position of the negative-sense strand primer-binding site also differs between caulimoviruses and soymoviruses, as does the presence of an intergenic region between ORF5 and 6.
Virion
Morphology
Isometric virions are 52 nm in diameter with an icosahedral T7 symmetry. The virion capsid consists of three concentric shells, built from 420 P4 proteins assembled as 60 hexavalent and 12 pentavalent capsomers. The inner cavity has a diameter of about 25 nm. P3 proteins (virion-associated protein, VAP) are incorporated in the virion as a triskelion structure that cements three hexavalent or pentavalent capsomers together. The N-terminus of the P3 protein is facing out of the cauliflower mosaic virus (CaMV) capsid. It contains two coiled-coil motifs with opposite handedness, forming dimers by anti-parallel interaction with adjacent P3 molecules to create a network around the virus particle (Plisson et al., 2005). The C-terminus of the P3 protein, which has a nucleic acid-binding motif, is embedded inside the pores surrounding the capsomers and traverses the P4 protein layers to reach the genomic DNA.
Through the same coiled-coil domains, virions interact with either the aphid transmission factor (ATF) or the movement protein (MP), both hosting coiled-coil domains at their C-terminus, to mediate aphid transmission and movement function, respectively (Leh et al., 1999, Stavolone et al., 2005).
Physicochemical and physical properties
Virions have a buoyant density of 1.35–1.38 g cm−3 in CsCl. Sedimentation coefficients are between 215 and 245 S.
Nucleic acid
Virions contain a single molecule of non-covalently closed circular dsDNA of 7.8–8.2 kbp. The negative-sense strand DNA has a single discontinuity and the positive-sense strand DNA, two or three discontinuities.
Proteins
Virions have two proteins, the capsid protein (CP, P4) and the VAP (P3). The VAP decorates the surface of the virion, facilitates interaction of the virion with host and other viral proteins [i.e. the movement protein (P1) and the aphid transmission factor (P2)], and is not essential for virion formation. Three domains are recognized in the CP: the acidic, N-terminal intrinsically disordered (NID) domain, the capsid (CA) domain and the nucleocapsid (NC) domain. The NC domain contains a zinc finger knuckle at the C-terminus and a basic, intrinsically disordered region at the N-terminus. Three isoforms of the CaMV CP are observed (44, 39 and 37 kDa). The N-terminus of the 44 kDa protein arises from aspartic protease cleavage of the CP precursor (protein P4) at alanine 77; the C-terminus lies between positions 435 and 440. There is a single PEST degradation signal in the NID domain of the 44 kDa protein, and targeted degradation of this domain allows nuclear targeting of mature virions, facilitated by a nuclear localization signal (RKRK) at positions 122–125. Three serine residues in the NID domain of the 44 kDa protein are phosphorylation targets, and phosphorylation enhances targeted degradation.
The P1 protein belongs to the 30K superfamily of viral movement proteins. It uses the endocytic pathway to target plasmodesmata where it forms tubular structures that replace the desmotubule of plasmodesmata and increases the size exclusion limit. Electron microscopic studies suggest that whole virions traffic between the cells utilizing the tubules.
The P2 protein is an aphid transmission helper component (Uzest et al., 2007) and along with the P3 protein, forms electron lucent transmission bodies (TBs) that occur individually in infected cells. These TBs are ingested by the aphid vector and, in an interaction involving the N-terminus of the P2 protein, bind to the tip of the aphid stylet (Plisson et al., 2005, Hoh et al., 2010, Webster et al., 2018). The P3 protein is then released and the bound P2 protein then binds P3-decorated virions during subsequent intracellular probes by the aphid.
The P6 protein (transactivator (TA)/viroplasm protein) is multifunctional and is the major component of the electron-dense inclusion bodies (EDIBs), the so-called ‘virion factories’ (Schoelz and Leisner 2017). P6 acts as a translational transactivator, allowing re-initiation of translation of downstream ORFs in the polycistronic 35S RNA. The P6 protein forms motile inclusion bodies that traffic along actin microfilaments, delivering virions to the plasmodesmata (Schoelz et al., 2016). Finally, the P6 protein is a symptom determinant and suppresses RNA silencing by modifying DICER activity, as well as inhibiting salicylic acid-dependent plant defence responses (Pooggin and Ryabova 2018).
Carbohydrates
The N-terminal region of the CaMV CP that is surface-exposed on the virion is glycosylated.
Genome organization and replication
The genome contains seven ORFs and large and small intergenic regions (Figure 1.Caulimovirus). The seventh ORF (ORF7) is present in all caulimoviruses downstream of the pgRNA leader and its translation can be initiated by ribosome shunting (Pooggin et al., 1999), but a corresponding protein has not yet been found in infected plants. The large intergenic region, containing the pregenomic RNA (35S) promoter, the RNA polyadenylation signal and the negative-sense strand primer-binding site, is located between ORF6 and ORF1. Electron-dense inclusion bodies (the viroplasm), which are composed primarily of P6, occur in the cytoplasm and are the site of viral DNA and protein synthesis, morphogenesis and storage of virions.
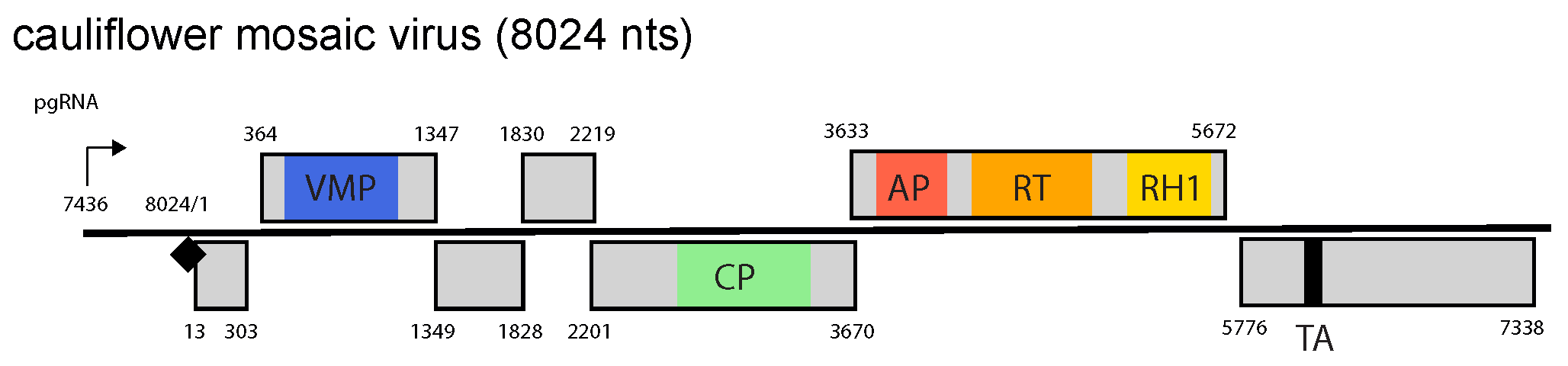 |
|
Figure 1. Caulimovirus. Caulimovirus genome organization. The linearized map begins at the pgRNA transcription start site (black arrow, mapped or predicted ca. 32 nts downstream of TATA box; see (Pooggin et al., 1999) and references therein). The numbering begins from the first nucleotide of the Met-tRNA primer binding site (black diamond). Light grey boxes mark open reading frames (ORFs). Conserved protein domains as listed in the Pfam database (http://xfam.org) are colored: blue is the viral movement protein (VMP) (PF01107), red is the retropepsin (pepsin-like aspartic protease) (AP) (CD00303), orange is the reverse transcriptase (RT) (CD01647) and yellow is the RNase H1 (RH1) (CD06222). The conserved C-terminus of the coat protein (CP) is marked green. The conserved translation transactivator (TA) domain is shown in black. |
Two major capped and polyadenylated transcripts (35S and 19S RNAs) are produced. The 35S RNA serves as the template for reverse transcription and as a polycistronic mRNA for proteins P1 to P5. P3, P4 and P5 can be translated from a spliced pgRNA (Pooggin and Ryabova 2018). P6 is translated from the 19S RNA and enables re-initiation of translation of downstream ORFs in a polycistronic pgRNA (and its spliced versions) after stop codons are passed. Several spliced versions of the 35S RNA are generated, some containing ORF3 and downstream sequences and others, ORF1 and 2 fused in-frame. This splicing is thought to prevent excessive expression of P2, which is inhibitory to virus replication. As much as 70% of the total viral RNA population is spliced. The caulimovirus CaMV also produces an 8S RNA from the pgRNA leader region, which serves as a decoy, diverting the antiviral RNA silencing machinery from other regions of the viral genome (Blevins et al., 2011). Such a decoy strategy of silencing evasion is also employed by the tungrovirus rice tungro bacilliform virus (Rajeswaran et al., 2014) and presumably members of other (but not all) genera of Caulimoviridae (Pooggin and Ryabova 2018).
Biology
Caulimoviruses are transmitted in a semi-persistent manner by aphids; Lamium leaf distortion virus (LLDV) has no known vector. Transmission requires a virus-encoded protein (aphid transmission factor) that is encoded by ORF2 (Uzest et al., 2007). All caulimoviruses except strawberry vein banding virus and LLDV are mechanically transmissible. Seed transmission is not recorded.
Natural host ranges are narrow (restricted to a single plant family) although some caulimoviruses have been experimentally transmitted to hosts in two to four plant families. Hosts are limited to dicotyledonous plants.
Antigenicity
Caulimoviruses are moderately to strongly immunogenic. Serological cross-reactivity between the isolates of different species is moderate to strong.
Derivation of names
Caulimovirus: derived from cauliflower mosaic virus, member of the type species of the genus.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Host range
- Differences in polymerase (RT + RNAse H) nt sequences of more than 20%
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
Aquilegia necrotic mosaic virus |
ANMV |
|
|
Dahlia common mosaic virus |
DCMV |
|
|
Eupatorium vein clearing virus |
EU569831 (NC_010738) |
EVCV |
|
Plantago virus 4 |
PlV-4 |
|
|
Sonchus mottle virus |
SMoV |

