Family: Parvoviridae
Susan F. Cotmore, Mavis Agbandje-McKenna, Marta Canuti, John A. Chiorini, Anna-Maria Eis-Hubinger, Joseph Hughes, Mario Mietzsch, Sejal Modha, Mylène Ogliastro, Judit J. Pénzes, David J. Pintel, Jianming Qiu, Maria Soderlund-Venermo, Peter Tattersall and Peter Tijssen
The citation for this ICTV Report chapter is the summary published as Cotmore et al., (2019):
ICTV Virus Taxonomy Profile: Parvoviridae, Journal of General Virology, 100: 367–368.
Corresponding author: Susan F. Cotmore ([email protected])
Edited by: Balázs Harrach and Stuart G. Siddell
Posted: December 2018
PDF: ICTV_Parvoviridae.pdf
Summary
Members of family Parvoviridae are small, resilient, non-enveloped viruses with linear, single-stranded DNA genomes of 4–6 kb (Table 1. Parvoviridae). Viruses in two subfamilies, the Parvovirinae and Densovirinae, are distinguished primarily by their respective ability to infect vertebrates (including humans) versus invertebrates. Being genetically limited, most parvoviruses require actively dividing host cells and are host and/or tissue specific. Some cause diseases, which range from sub-clinical to lethal. A few require co-infection with helper viruses from other families.
Table 1. Parvoviridae. Characteristics of members of the family Parvoviridae.
| Characteristic | Description |
| Typical member | human parvovirus B19-J35 (AY386330), species Erythroparvovirus primate1, subfamily Parvovirinae |
| Virion | Small, non-enveloped, T=1 icosahedra, 23–28 nm in diameter |
| Genome | Linear, single stranded DNA of 4–6 kb with short terminal hairpins |
| Replication | Rolling hairpin replication, a linear adaptation of rolling circle replication. Dynamic hairpin telomeres prime complementary strand and duplex strand-displacement synthesis; high mutation and recombination rates |
| Translation | Capped mRNAs; colinear ORFs accessed by alternative splicing, non-consensus initiation or leaky scanning |
| Host range | Parvovirinae: vertebrates, currently mammals, birds, reptiles. Densovirinae: invertebrates, currently insects, crustacea, echinoderms |
| Taxonomy | Realm Monodnaviria, kingdom Shotokuvirae, phylum Cossaviricota, class Quintoviricetes, order Piccovirales: 3 subfamilies, Parvovirinae, Hamaparvovirinae and Densovirinae; 28 genera, 243 species |
Virion
Morphology
Virions are small, non-enveloped particles, 23–28nm in diameter, with no known essential lipids, carbohydrates, accessory proteins or histones. They are exceptionally rugged, often remaining infectious in the environment for months or years. X-ray crystallographic and cryoEM image reconstruction studies reveal a highly characteristic particle structure with T=1 icosahedral symmetry (Agbandje-McKenna and Kleinschmidt 2011, Halder et al., 2013). Capsids are assembled from sixty structurally-equivalent polypeptide chains derived from the C-terminal end of a single capsid viral protein (VP) sequence. These monomers interdigitate extensively to create 60 asymmetric units per particle, which co-ordinate at twelve 5-fold, twenty 3-fold and thirty 2-fold axes (Figure 1. Parvoviridae). Typical capsid surface features include depressions at each icosahedral two-fold axis, elevated protrusions surrounding each 3-fold axis, and raised cylindrical projections encircling each 5-fold vertex that are themselves surrounded by extensive canyon-like depressions. Such surface features are conspicuous in most family members, but their relative elevation varies between subfamilies and genera (Figure 1. Parvoviridae). Each 5-fold cylinder potentially encloses a central channel, which penetrates from the outer surface of the capsid to the particle interior. These serve as portals, mediating the entry and eventual exit of the viral genome from intact particles and in members of some genera, the phased extrusion of N-terminal regions from certain VPs that carry functional motifs required at specific stages in the viral life cycle (Plevka et al., 2011, Cotmore and Tattersall 2014, Meng et al., 2013). Some pathogenic shrimp and mosquito viruses (in genera Penstyldensovirus and Brevidensovirus respectively) have VP proteins that are relatively short and the loops connecting their beta-barrel strands appear minimal, resulting in particles with small diameters and a relatively smooth appearance (Figures 1. Parvoviridae and 2. Parvoviridae)(Kaufmann et al., 2010, Tijssen et al., 2016). To date, structures of the VP1 N-terminal unique region have not been resolved at the atomic level, likely because their low copy number and/or flexibility makes them undetectable after rotational averaging in the context of the capsid. However, circular dichroism of adeno-associated virus 6 (AAV6) virus-like particles (VLPs) indicates that at neutral pH these regions adopt an α-helical conformation and unfold reversibly in a pH-dependent manner as the pH is lowered from 7.5 to 4.0 (Venkatakrishnan et al., 2013). Around 20 nucleotides from the 5′-end of the viral genome may remain exposed at the surface of newly released virions, bearing a covalently-attached copy of the viral replication initiator protein, NS1, at its 5′-end (Cotmore and Tattersall 1989, Prasad and Trempe 1995). These are vestiges of the genome excision and packaging mechanisms (Cotmore and Tattersall 2014, King et al., 2001), and are vulnerable to enzymatic removal in the environment without affecting subsequent particle infectivity.
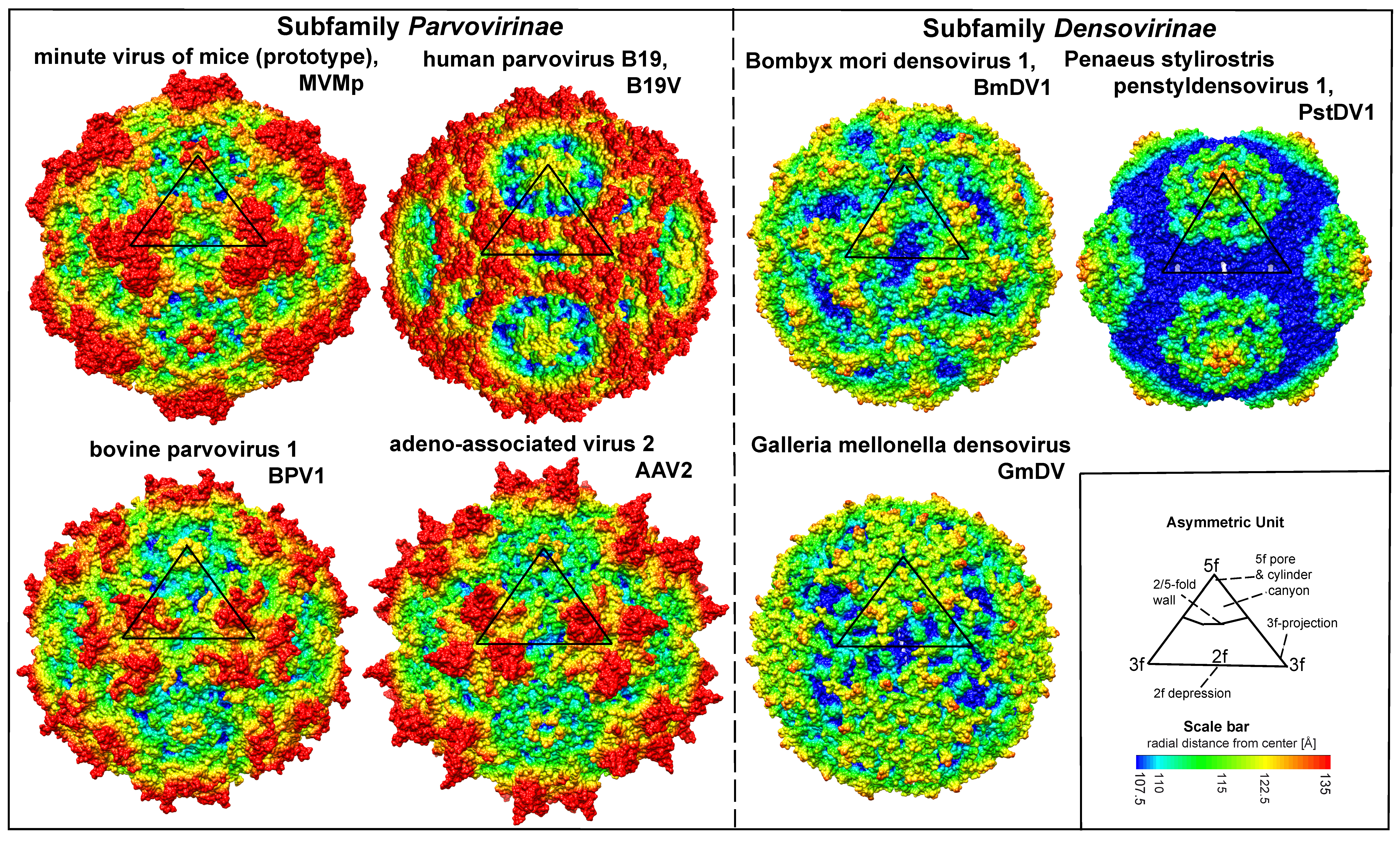 |
| Figure 1. Parvoviridae. Surface images showing capsid topologies of viruses from selected genera in subfamilies Parvovirinae and Densovirinae. Radial distances from the particle centre are colour coded (blue to red), as indicated on scale bar. An asymmetric unit diagram is labelled to indicate icosahedral 2-, 3- and 5- fold axes and the 2/5-fold wall. The position of a single asymmetric unit is indicated on each model. Atomic co-ordinates were obtained as follows: minute virus of mice (prototype) (Protoparvovirus, PDB ID 1Z14) (Kontou et al., 2005, Agbandje-McKenna et al., 1998), human parvovirus B19 (Erythroparvovirus, PDB ID 1S58) (Kaufmann et al., 2004), bovine parvovirus 1 (Bocaparvovirus, PDB 4QC8) (Kailasan et al., 2015b), adeno-associated virus 2 (Dependoparvovirus, PDB 1LP3) (Wigfield and Eatock 1990), Bombyx mori densovirus 1 (Iteradensovirus, PDB ID 3POS) (Kaufmann et al., 2011), Penaeus stylirostris penstyldensovirus 1 (Penstyldensovirus, PDB 3N7X) (Kaufmann et al., 2010), Galleria mellonella densovirus (Ambidensovirus, PDB 1DNV) (Simpson et al., 1998). Figures were generated using the program Chimera (Pettersen et al., 2004). |
Each VP chain forms a core beta-barrel structure of at least 8 strands (labelled βB–βI in Figure 2. Parvoviridae) arranged as two adjacent beta sheets (that spell CHEF and BIDG, see Figure 2E. Parvoviridae), with the BIDG sheet forming the inner surface of the capsid shell. Individual beta strands are linked by loops of variable length, sequence, and conformation, most of which project towards the outer surface of the capsid and give individual viruses their unique surface topology. The apices of these loops are the most varied and defined as variable regions (VRs). Although these monomer structures can differ widely between members of different genera (cf. Figure 2. Parvoviridae A–D), for viruses within a genus they are typically highly conserved. This is illustrated for members of genus Protoparvovirus in Figure 2E. Parvoviridae, where the Cα positions of the core secondary structure elements (β sheets A–I and helix A) of viruses from 4 different species are completely superimposed, and even the variable loops between these conserved structures follow very similar topologies (Figure 2E. Parvoviridae) (Ilyas et al., 2018). Thus in general, parvovirus capsid structures are significantly more conserved than their protein sequence identities suggest: for example, the capsid amino acids of minute virus of mice (prototype, MVMp) and porcine parvovirus (PPV) share around 52% sequence identity, but 97% of their alpha carbons occupy similar positions (Figure 2E. Parvoviridae), and even between PPV and the insect virus Galleria mellonella densovirus (GmDV, genus Ambidensovirus; cf. Figure 2. Parvoviridae C and E), 40% of capsid residues align despite sharing only 9% sequence identity. Accordingly, capsid structures serve as a strong indicator of parvovirus phylogenetic identity.
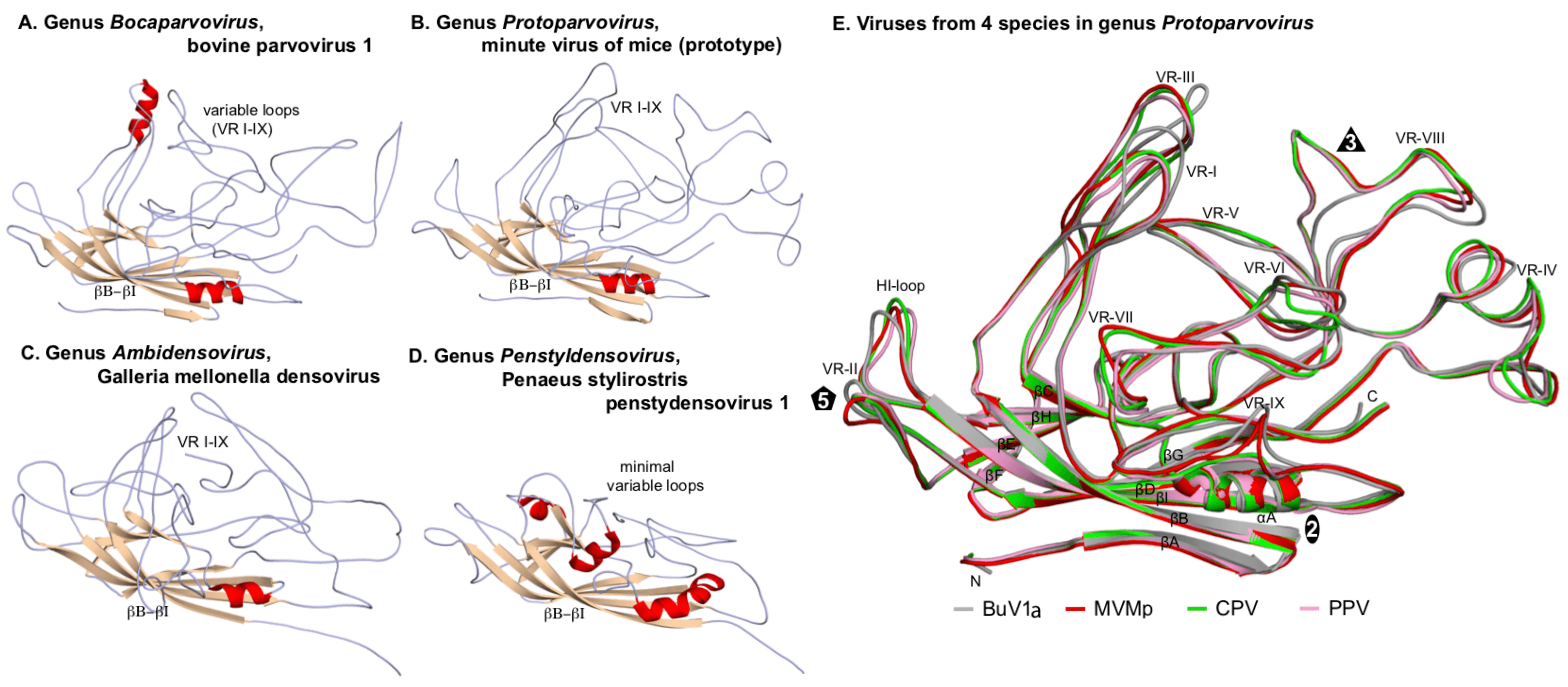 |
| Figure 2. Parvoviridae. Folding of individual VPs in the parvovirus capsid shell. Capsid monomers from viruses in subfamilies Parvovirinae (A, B) and Densovirinae (C, D) are shown as a series of gray coils with β-sheets in brown and α-helices in red. β-barrels (βB–βI) and variable loops (VR I–IX) are indicated. (E) Superposed capsid protein monomers from viruses in 4 species of genus Protoparvovirus. Colour coded VP2 monomers from bufavirus 1a (human) (BuV1a; gray), minute virus of mice (prototype) (MVMp; red), canine parvovirus (CPV; green) and porcine parvovirus (PPV; pink) are overlaid, highlighting their conserved folds. The β-strands (βA–βI), VR regions (VR-I to VR-IX), and the HI-loop are shown and labeled. Icosahedral 2-, 3-, and 5-fold axes are represented by an oval, triangle, and pentagon, respectively. Atomic co-ordinates were as indicated in Figure 1. Parvoviridae, with the addition of PPV (PDB 1K3V) (Simpson et al., 2002), CPV (PDB 2CAS) (Wu et al., 1993) and BuV1a (PDB 6BWX) (Ilyas et al., 2018). Images were generated using PyMOL (Schrodinger 2015). |
One consistent structural difference between virus capsids in members of the subfamilies Parvovirinae and Densovirinae involves the βA strand: whereas in vertebrate parvoviruses (Figure 2. . Parvoviridae A–B and E) βA folds back and hydrogen bonds with βB from its own subunit, in densoviruses βB is a linear extension of the βA strand (Figure 2. . Parvoviridae C–D), which allows βA to hydrogen bond with βB in the neighboring 2-fold related subunit, potentially enhancing the structural stability of this region. This is referred to as "domain-swapping".
Physicochemical and physical properties
Infectious particles comprise around 75% protein and 25% DNA, giving them buoyant densities of 1.39–1.43 g cm−3 in CsCl and relatively low sedimentation co-efficients (S20w) of 110–122S. Many current purification protocols rely on density gradient centrifugation through isotonic iodixanol gradients, where virions band at around 1.3 g cm−3 (Zolotukhin et al., 1999). Mature virions can withstand elevated temperatures (e.g. <55°C for 60 min), particularly in crude tissue extracts, but when purified they may be metastable, undergoing conformational shifts after incubation at 60–65°C that render them non-infectious (Cotmore and Tattersall 2014, Hulks et al., 1990). Virions generally tolerate exposure to pH 3–9 or lipid solvents, and are resistant to many disinfectants, but sensitive to UV irradiation.
Nucleic acid
The genome is a linear non-permuted ssDNA molecule of 4 to 6 kb in which a long coding region is bracketed by short (116 to ~550 nucleotide) imperfect palindromes that fold into dynamic hairpin telomeres (Figure 3. Parvoviridae). These hairpin sequences provide most of the cis-acting information required for DNA replication and packaging, and give rise to viral replication origins at the right- and left-ends of the genome when amplified through duplex replicative form (RF) DNA intermediates. Some parvoviruses preferentially excise and encapsidate ssDNA of negative polarity (e.g. MVM, genus Protoparvovirus), while others encapsidate strands of either polarity in equivalent (e.g. adeno-associated virus 2 (AAV2), genus Dependoparvovirus) or different proportions (e.g. bovine parvovirus 1 (BPV1), genus Bocaparvovirus). These strand preferences largely reflect the efficiency with which single-strands of each sense are displaced from RF DNA during replication, which in turn reflects the relative efficiency of the viral origins associated with the two genomic termini (Cotmore and Tattersall 2005b).
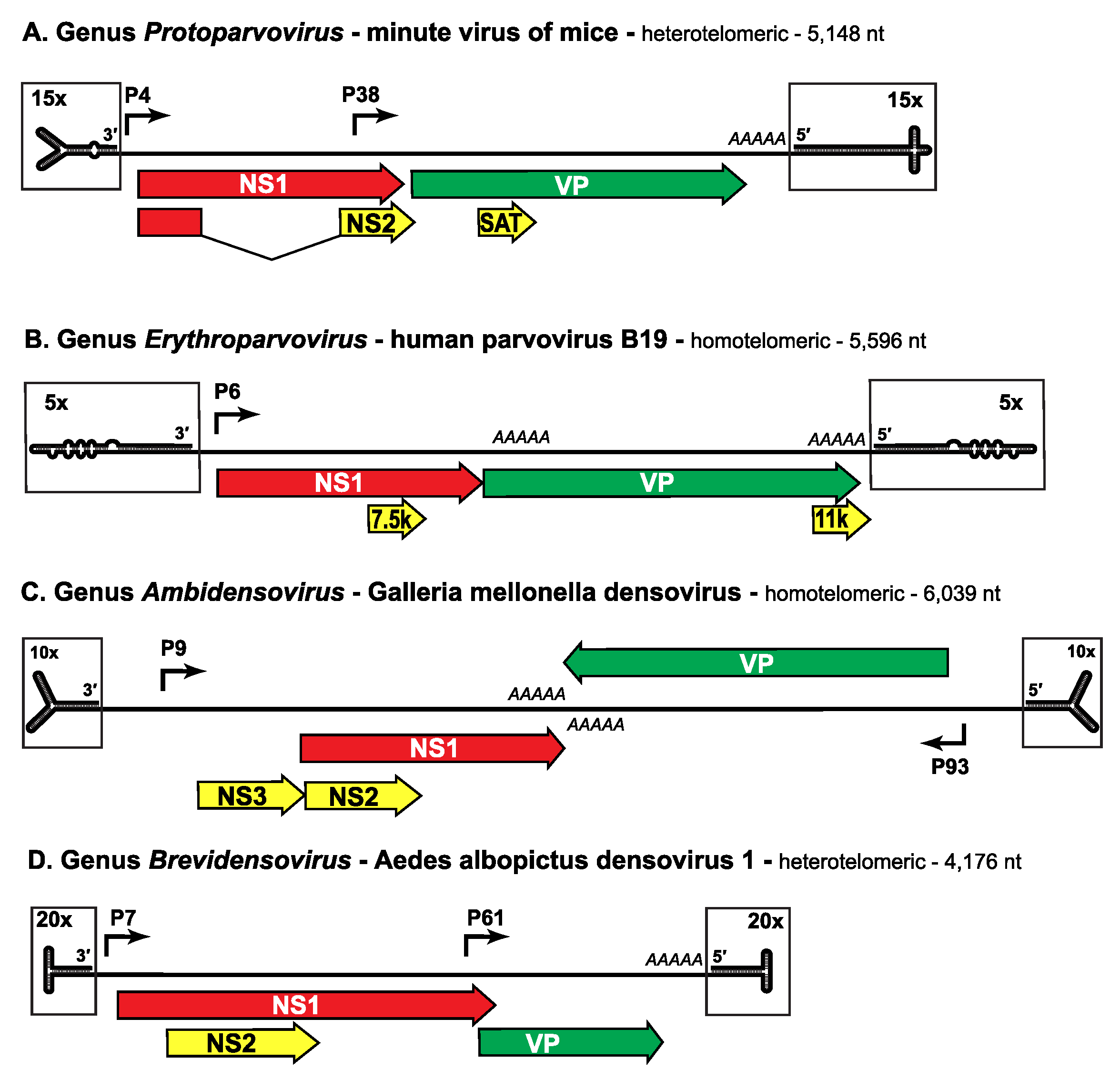 |
| Figure 3. Parvoviridae. Genetic strategies of representative viruses from family Parvoviridae. |
Negative-sense genomes of viruses from representative genera in subfamilies Parvovirinae (A, B) and Densovirinae (C, D) are shown as single lines terminating in boxed hairpin structures, which are magnified relative to the rest of the genome, as indicated. Hairpin structures are drawn to represent their lowest predicted free energy states. Major open reading frames that encode proteins are shown as arrowed boxes, some of which are linked by splicing to encode ancillary proteins. Regions encoding the replication initiator protein, NS1, are shaded red, those encoding the capsid proteins, VP, are green, and those unique to ancillary proteins are yellow (but these may also share common introns with NS1, as indicated). Solid arrows represent transcriptional promoters, AAAAA indicates polyadenylation sites, and SAT stands for “small alternatively translated protein”. Data was obtained from the following sources: minute virus of mice (Pintel et al., 1983), human parvovirus B19 (Liu et al., 2004), Galleria mellonella densovirus (Tijssen et al., 2003) and Aedes albopictus densovirus 1 (Boublik et al., 1994).
As listed in Tables 2A and B, viruses in some genera of both subfamilies are homotelomeric, meaning that their left- and right-end hairpin termini are similar and form part of larger terminal repeat (TR) sequences, whereas in other genera viruses are heterotelomeric, with disparate hairpins at the two ends of the genome that are processed during replication by a more conservative mechanism. Within a genus all viruses have very similar terminal hairpins, but these structures vary markedly in sequence and potential secondary structure between members of different genera (Figure 3. Parvoviridae), making them powerful indicators of phylogenetic identity. However, when cloned and amplified in bacterial plasmids or by polymerase-based methods such as PCR, many are prone to deletion and/or re-arrangement, rendering the genome non-infectious and limiting their use as diagnostic features in sequence-based taxonomy.
Table 2A. Parvoviridae. Virus characteristics in genera from subfamily Parvovirinae.
Genus and type species Exemplar virus of type species and other well known viruses in the genus, with abbreviations | Spd
| DNA (kb)
| Genomic termini: homo- or hetero-telomeric size of TRs and/or HPs | Pol-IIa
| PLA2 motif in VP1
| APsb
| Commentsc
|
Amdoparvovirus Carnivore amdoparvovirus 1 Aleutian mink disease parvovirus, ADV | 4 | ~4.8 | hetero-telomeric; left ~116 nt right ~240 nt | 1 | No | NS2, NS3 | Some VI; Some MTG; Persistent infections |
Aveparvovirus Galliform aveparvovirus 1 turkey parvovirus, TuPV | 1 | ~5.3
| homo-telomeric; TRs ~206 nt HP ~39 nt | 2 | No | NP | |
Bocaparvovirus Ungulate bocaparvovirus 1 bovine parvovirus 1, BPV1; human bocavirus 1, HBoV1; minute virus of canines, MVC | 21 | ~5.5 | hetero-telomeric; left ~140–180 nt right ~120–200 nt | 1* | Yes | NP1, NS2, NS3, NS4 | 3 VI; many MTG; HU |
Copiparvovirus Ungulate copiparvovirus 1 bovine parvovirus 2, BPV2; porcine parvovirus 4, PPV4 | 2 | ~5.6 | Unknown, probably homo-telomeric | U‡ | Yes | U‡ | MTG |
Dependoparvovirus Adeno-associated dependoparvovirus A primate adeno-associated viruses, AAV1–4, 6–12; adeno-associated virus 5, AAV5; Muscovy duck parvovirus, DPV | 7 | ~4.7 | homo-telomeric; TRs ~145–454 nt HP ~125–415 nt | 3 | Yes | Rep 40, AAP | Many VI; Some MTG; HU; AAVs HV; DPVs helper virus independent |
Erythroparvovirus Primate erythroparvovirus 1 human parvovirus B19, B19V | 6 | ~5.6 | homo-telomeric; TRs ~383 nt HP ~365 nt | 1 | Yes | 11k, 7.5k | 4 VI; 2 MTG; HU |
Protoparvovirus Rodent protoparvovirus 1 minute virus of mice, MVM; bufavirus 1a (human), BuV1a; canine parvovirus, CPV; feline parvovirus, FPV; porcine parvovirus, PPV; parvovirus H1, H1 | 11 | ~5.1 | hetero-telomeric; left ~140–180 nt right ~180–200 nt | 2 | Yes | NS2, SAT | Many VI; Some MTG; HU |
Tetraparvovirus Primate tetraparvovirus 1 human parvovirus 4, PARV4 | 6 | ~5.3 | Unknown | 2 | Yes | NS2 | MTG; HU |
a: Pol-II: Pol-II promoters
b: APs: Ancillary Proteins
c: Comments: VI = Virus isolated; MTG = Metagenomic; HU = Human host; HV = Helper virus
d: Sp: species
*: HBoV1 also has 1 pol-III
‡: U = Unknown
Table 2B. Parvoviridae. Virus characteristics in genera from subfamily Densovirinae.
Genus and type species Exemplar virus of type species and other well known viruses in the genus, with abbreviations | Spd
| DNA (kb)
| Genomic termini: homo- or hetero-telomeric size of TRs and/or HPs | Pol-IIa
| PLA2 motif in VP1
| APsb
| Commentsc
|
Ambidensovirus Lepidopteran ambidensovirus 1 Galleria mellonella densovirus, GmDV; sea star-associated densovirus, SSaDV | 11 | ~6 | homo-telomeric; TRs ~550 nt HPs ~139 nt |
2–3 |
Yes |
NS2, NS3 | Most VI; Spliced and unspliced RNA |
Brevidensovirus Dipteran brevidensovirus 1 Anopheles gambiae densovirus, AgDV; Aedes albopictus densovirus 2, AalDV2 | 2 | ~4.2 | hetero-telomeric; left ~135 nt right ~180 nt | 3* | No | NS2 | Most VI; Unspliced transcripts |
Hepandensovirus Decapod densovirus 1 Fenneropenaeus chinensis hepandensovirus, FcHDV; Penaeus monodon hepandensovirus 1, PmoHDV1 | 1 | ~6.3 | hetero-telomeric; HPs ~0.2kb | 3 | No | NS2 | Most MTG; Unspliced transcripts |
Iteradensovirus Lepidopteran iteradensovirus 1 Bombyx mori densovirus 1, BmDV1 | 5 | ~5 | homo-telomeric; TRs ~250 nt HPs ~165 nt | 2 | Yes | NS2 | Most VI; Unspliced transcripts |
Penstyldensovirus Decapod penstyldensovirus 1 Penaeus stylirostris penstyldensovirus 1, PstDV1 | 1 | ~4 | Unknown | 3 | No | NS2 | Most MTG; Unspliced transcripts |
unassigned Orthopteran densovirus 1 Acheta domestica mini ambidensovirus, AdMADV | 1 | ~4.9 | homo-telomeric; TRs ~199 nt HPs ~113 nt | 2 | Yes | NS2, NS3 | VI |
a: Pol-II: Pol-II promoters
b: APs: Ancillary Proteins
c: Comments: VI = Virus isolated; MTG = Metagenomic
d: Sp: species
*: two overlap
Proteins
Capsids are assembled from a nested set of sixty VPs, typically encoded on a single structural gene that includes the entire coding sequence for VP1 (generally 75–100kDa), while one or more smaller forms (VP2–5) share a common C-terminal sequence but have N-terminal truncations of different length. Exceptions to this strict pattern are found in some viruses from genus Ambidensovirus, (see genus section). The VP1-specific region of viruses in all genera except Amdoparvovirus, Aveparvovirus, and Brevidensovirus contain an atypical, broad-specificity phospholipase A2 (PLA2) domain with characteristic protein motifs that correspond to a calcium binding loop (GPGN) and the PLA2 active-site (DxxAxxHDxxY) (Zádori et al., 2001, Canaan et al., 2004). Typically there are 5–10 copies of the VP1 per capsid.
Lipids and Carbohydrates
Virions lack essential lipids. No viral proteins are known to be glycosylated.
Genome organization and replication
Viruses have 2 major gene cassettes; a non-structural replication initiator gene (NS) located in the 3′ (by convention the "left") half of the negative-sense strand, and a single capsid sequence (VP) located in the right half. (Figure 3. Parvoviridae). The replication initiator protein (NS1, sometimes called the replicase) has an endonuclease domain that combines sequence-specific duplex DNA-binding and site-specific single-strand nicking activity, followed by a superfamily 3 DNA helicase domain, but it does not have polymerase activity, and these viruses rely entirely upon host DNA polymerase(s) to amplify their genomes during productive infection. Most genomes also encode a small number of ancillary proteins in alternate and/or overlapping open reading frames, thus maximally exploiting the available DNA. These ancillary proteins are commonly genus specific and play many different roles in various aspects of the viral life cycle, although only the unique NP1 proteins encoded by members of genus Bocaparvovirus are known to influence RNA processing events by suppressing internal polyadenylation and splicing of an upstream intron (King et al., 2001, Choi et al., 2005, Mihaylov et al., 2014, Brockhaus et al., 1996, Eichwald et al., 2002, Engelsma et al., 2008, Miller and Pintel 2002, Sonntag et al., 2010, Earley et al., 2017, Sukhu et al., 2013, Fasina et al., 2016, Fasina et al., 2017, Zádori et al., 2005, Mészáros et al., 2017b, Zou et al., 2016, Chen et al., 2010b). Expression strategies vary substantially between, and sometimes even within, genera, as indicated in Figure 3. Parvoviridae and in the genus sections of this report, but most vertebrate parvoviruses encode NS and VP proteins in tandem on a single DNA strand. However, some densoviruses are "ambisense", meaning that transcription of the NS and VP genes proceeds from promoters at the ends of each complementary strand (Figure 3. Parvoviridae). Viruses in some genera use a single transcriptional promoter near the end of their coding sequences, and the regulation of protein expression is post-transcriptional, whereas in other genera, viruses use two or three promoters, on the same or different template DNAs, as detailed in the genus sections of this report, with NS1 commonly taking on the additional function of phasing/modulating promoter activity. When proteins are encoded in multiple co-linear frames, alternative splicing, suboptimal translational initiation or leaky scanning mechanisms are used to create different forms of the gene products. Messenger RNAs are capped and polyadenylated. Human bocavirus 1 (HBoV1) also expresses BocaSR, a small non-coding RNA polymerase-III transcript that resembles primate adenovirus ”virus associated” VA1 RNA in both nucleotide sequence and secondary structure. This RNA regulates expression of some NS proteins and is essential for viral DNA replication (Wang et al., 2017b).
Parvovirus genomes remain protected within the virion until after they are trafficked into the host cell nucleus (Sonntag et al., 2006, Mäntylä et al., 2017, Ros et al., 2017, Vendeville et al., 2009), but how they are uncoated and how their hairpins and single-stranded DNAs are protected from inactivation by cellular repair networks remains uncertain. Since they need to co-opt the cellular DNA replication machinery, host cells must generally enter S-phase before viral DNA synthesis can initiate. Viral replication proceeds via a “rolling hairpin” mechanism (Tattersall and Ward 1976), in which unidirectional strand displacement synthesis is coupled with the sequential unfolding and refolding of the hinge-like hairpin termini, to allow the synthesis of continuous duplex replicative form (RF) DNA intermediates (Figure 4. Parvoviridae). Progeny single-strands are then excised from these intermediates by the viral replication initiator protein (Figure 4. Parvoviridae) (Cotmore and Tattersall 2014, Snyder et al., 1990, Berns and Parrish 2013). These proteins (generally called NS1, but Rep1 or Rep68/78 in genus Dependoparvovirus) are multi-functional, typically of around 70–100 kDa in mass, with an N-terminal site- and strand-specific HuH-family endonuclease domain, which also has site-specific duplex DNA-binding activity (Cotmore and Tattersall 2014, Snyder et al., 1990, Berns and Parrish 2013, Hickman et al., 2004, Tewary et al., 2013, Tewary et al., 2015, Im and Muzyczka 1990). This is linked to a central superfamily 3 (SF3) helicase domain that has 3′-to-5′ processivity (Im and Muzyczka 1990, James et al., 2003), and a C-terminal region with transcriptional activation and/or other activities that differ between genera. NS1 mediates the excision of both ends of the genome from duplex replication intermediates by carrying out a trans-esterification reaction that introduces a site-specific single-strand nick into specific duplex origin sequences. These nicks liberate a 3′ nucleotide, which can then prime new rounds of replication, but NS1 is left covalently attached to the new 5′-end of the DNA via its active site tyrosine. In consequence, a copy of NS1 remains covalently attached to the 5′-end of all RF and progeny viral DNA molecules, where it may persist throughout replication, packaging, and virion release (Cotmore and Tattersall 2003).
Figure 4. Parvoviridae illustrates how the unfolding and refolding of the terminal hairpins of a heterotelomeric virus, such as MVM, generates monomeric and concatemeric duplex intermediates. In step a, the base-paired 3′-nucleotide of the 3′-hairpin primes conversion of virion DNA to the first duplex intermediate. This generates a monomer-length duplex molecule in which the two strands are covalently continuous at the viral 3′-telomere. This serves as the first viral transcription template, allowing expression of NS1. Before NS1 accumulates, replication cannot proceed beyond the position indicated in step a), and the 3′-end of the new strand may become ligated to the original 5′-end (step b), creating a covalently continuous duplex at both ends of the genome. NS1 resolves the right-end structure (and all subsequent covalently continuous right-hand ends) by carrying out a "hairpin transfer" reaction, in which it creates a site-specific single-strand nick in the new DNA (step c). With the help of NS1, a replication fork established at this nick now unfolds and copies the hairpin, replacing the original sequence with its inverted complement (step d). Parvoviral hairpins are imperfect palindromes, and since this inversion occurs with every round of replication, progeny genomes comprise equal numbers of each terminal orientation, dubbed “flip” and “flop”. In step (e), extended-form right-end termini are melted out and reformed into hairpin “rabbit ear” structures in a process that is again facilitated by direct binding of NS1. This allows the newly synthesized DNA to create the base-paired hairpin structures needed to prime synthesis of additional linear sequences (step f), giving rise to a palindromic duplex dimer intermediate (step g). The dimer can undergo the same right-end “rabbit-ear” rearrangement (step h), allowing synthesis of tetrameric concatemers (step h), in which alternating unit length genomes are fused in left-end:left-end and right-end:right-end orientations.
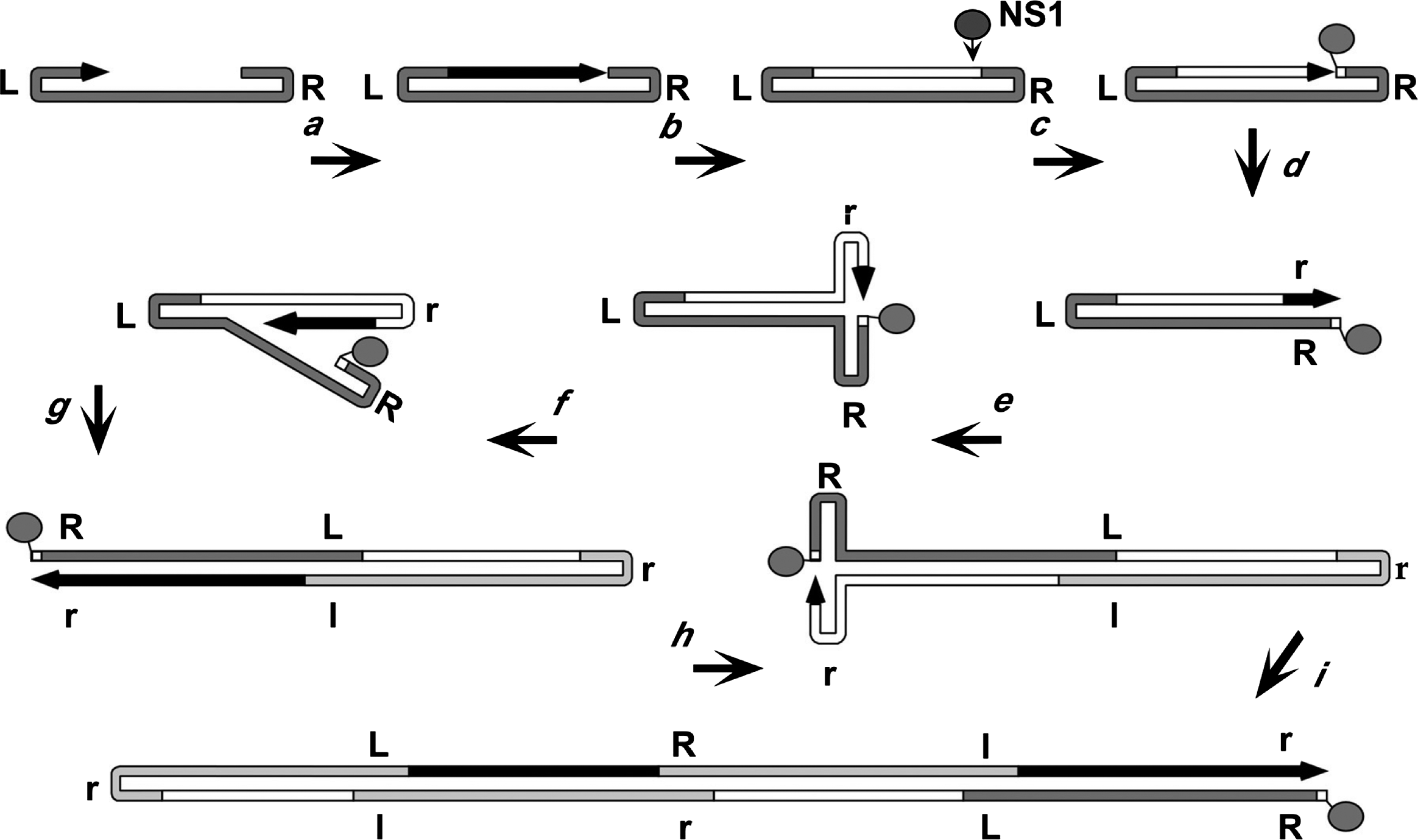 |
| Figure 4. Parvoviridae. Rolling hairpin synthesis, modelled for a heterotelomeric parvovirus. An arrowed black line represents the newly synthesized DNA of the growing 3′-end, with the original genome in dark grey. Light grey denotes progeny genomes embedded within the oligomeric intermediates. Upper and lower cases of R and L represent flip and flop forms of the right and left ends, respectively. |
During the replication of homotelomeric viruses, such as human parvovirus B19 (B19V) or AAV2, hairpin transfer reactions of the type shown in Figure 4. Parvoviridae are carried out at both genomic termini, but for heterotelomeric viruses hairpin transfer occurs only at the right-end of the genome, because different structural and co-factor requirements are needed to activate the NS1 nickase at the two disparate termini. For MVM, left-end telomeres are then excised from duplex left-end:left-end junctions in these concatemers by a conservative process called junction resolution which proceeds through a heterocruciform intermediate, liberating new termini in a single "flip" sequence orientation (Cotmore and Tattersall 2003). As preassembled empty viral particles accumulate, displaced progeny genomes are packaged into them via a portal at one of the icosahedral 5-fold axes in the capsid, driven by the NS1 3′-to-5′ helicase (Plevka et al., 2011, King et al., 2001, Yoon-Robarts et al., 2004). Progeny virions may be exported from cells (Maroto et al., 2004), or accumulate in the nucleus until released by cell lysis (Bär et al., 2008, Bär et al., 2013).
The products of parvoviral replication evoke profound DNA damage responses (DDR) in host cells, mediated by any or all of the three major phosphatidyl inositol 3 kinase-like-kinase pathways (ATM, ATR and DNA-pk), but details show extreme variation between viruses (Luo and Qiu 2013, Deng et al., 2016, Adeyemi and Pintel 2014, Fuller et al., 2017, Majumder et al., 2017). Individual parvoviruses exploit these altered environments in complex and distinctive ways, but in each case productive infection absolutely requires activation of at least one DDR pathway. This requirement in part reflects the profound effect damage responses have on cellular DNA replication, which typically terminates in S-phase and/or just prior to G2M, allowing the cell's resources to be diverted for viral replication. Although parvovirus DNA is amplified by a subset of the cellular replication machinery, unidirectional viral forks appear to evade DNA editing and repair functions that normally limit mutation rates in cellular DNA, and their single-stranded progeny genomes potentially conserve every mutation. Accordingly, parvovirus mutation rates are high, resulting in a single nucleotide substitution for approximately every 104 bases (López-Bueno et al., 2003, Shackelton et al., 2005, Shackelton and Holmes 2006, López-Bueno et al., 2008), which means that each time the 5 kb genome is copied, packaged and transmitted, approximately half of its progeny harbor a mutation. Recombination between replicating genomes is also rampant. As a result, diverse strains of viruses can be generated rapidly, which may both co-exist and recombine within a particular environment (Pearson and Pintel 2000, Canuti et al., 2016).
Biology
Parvoviruses enter cells using a wide variety of cell surface receptor molecules (Huang et al., 2014a, Allison et al., 2014, Pillay et al., 2017, Chiorini 2016), and are internalized by endocytic pathways. Virions are metastable (Cotmore and Tattersall 2014, Meng et al., 2013); in endosomes many undergo a conformational shift, exposing phospholipase A2 (PLA2) domains on their VP1 N-termini that are required for lipid bilayer penetration, although the exact mechanism of membrane disruption remains uncertain (Girod et al., 2002, Farr et al., 2005). Intracellular trafficking appears variable but ultimately intact virions are delivered into the cell nucleus (Sonntag et al., 2006, Mäntylä et al., 2017, Ros et al., 2017, Vendeville et al., 2009) where their genomes become exposed. In vitro studies of MVM indicate that uncoating can be promoted by calcium depletion, and occurs in a 3′-to-5′ direction through a pore at one of the icosahedral 5-fold axes, leaving the 5′-end of the DNA still firmly attached to the otherwise intact capsid (Cotmore et al., 2010, Cotmore and Tattersall 2012). For AAV2 there is evidence that transcription initiation is capsid associated (Aydemir et al., 2016).
Unlike most DNA viruses, parvoviruses cannot drive non-cycling cells into S phase, but must typically wait for cells to make the transition under their own cell cycle control. This makes actively dividing cell populations essential for productive infection, and renders foetal or neonatal animals particularly vulnerable because they have a wide range of actively dividing differentiated cell types. Many parvoviruses cross the placenta, and some may be teratogenic. Their inability to induce S-phase in host cells likely explains why parvoviruses are not oncogenic. Since many host restrictions are relaxed when cells undergo oncogenic transformation, viruses in some species, such as Rodent protoparvovirus 1, are preferentially oncolytic (Geletneky et al., 2015, Geletneky et al., 2017). Remarkably, HBoV1 is able to achieve significant levels of viral replication in terminally differentiated airway epithelial cells that cannot enter S-phase (Deng et al., 2016), by exploiting components of an evoked DNA damage response. In contrast, AAVs compensate for their genetic limitations by co-opting helper viruses to support their productive replication and it is the helper virus that evokes the necessary replicative environment. Adenoviruses or herpesviruses serve this helper function most commonly, although vaccinia, papilloma, and even HBoV1 can also supply effective help in some situations (Atchison et al., 1965, Schlehofer et al., 1986, Wang et al., 2017a). In recent years, the inert nature of helper-free AAV gene delivery, together with the durability and diverse tissue-specificities of these particles, has allowed their use in transducing a wide range of species and tissues in vivo without detectable toxicity, making them powerful gene transfer vehicles for research and clinical studies (Samulski and Muzyczka 2014).
Although most parvovirus infections are inefficient, requiring high particle-to-infectivity ratios, their virions are exceptionally rugged, potentially offering long-term protection for the genome. Accordingly, once released into the environment they may remain infectious for months or years. In humans, this is manifest by the significant proportion of babies that become infected with HBoV1 in the early months of life, developing their own antibody responses as soon as waning maternal IgG levels render them vulnerable to infection from their environment (Kantola et al., 2015). While parvoviruses can induce a broad range of pathology in humans (Qiu et al., 2017), in most people these are rarely life threatening. This is in sharp contrast to the carnivore-infecting viruses from genus Protoparvovirus, such as canine parvovirus (CPV), which replicates in dogs in intestinal and bone marrow cell populations that remain dividing throughout the animals’ lifespan, and where mortality rates can reach around 90% if untreated (Kailasan et al., 2015a). Many densoviruses are also highly pathogenic, sometimes posing severe economic or ecological threats, for example in shrimp aquaculture (Tijssen et al., 2016).
Antigenicity
Members of subfamily Parvovirinae elicit powerful polyclonal antibody-mediated immune responses during productive infection that typically serve to limit systemic viremias. These antibodies provide a relatively long-lived record of past infections, which can be quantified by binding to recombinant virus particles. Such assays have been used to explore the epidemiology of newly-discovered parvoviruses within human populations (Lahtinen et al., 2011, Kantola et al., 2015, Väisänen et al., 2017, Väisänen et al., 2018), and can help to evaluate the presumed hosts of isolates identified by metagenomic virus-discovery approaches from equivocal sources (Väisänen et al., 2018).
Binding sites for individual monoclonal antibodies have been mapped for viruses from several genera using mutagenesis or cryoEM image reconstruction (Hafenstein et al., 2009, Tseng et al., 2015, Kailasan et al., 2016). In most cases, these footprints map to two antigenically dominant regions of the capsid surface that involve the icosahedral 3-fold protrusions and the wall between the 2-fold and 5-fold axes (Figure 1.Parvoviridae).
Derivation of names
Parvo: from Latin parvus meaning small
Denso: from Latin densus meaning thick or compact
Amdo: from Aleutian mink disease
Ave: from Latin aves indicating bird hosts of founding members
Boca: from the bovine and canine hosts of founding viruses
Copi: from cow and pig hosts of founding viruses
Dependo: from English depend meaning to require help
Erythro: from Greek erythros meaning red, indicating relationship to blood
Proto: from Greek protos meaning first
Tetra: from Greek tetra- meaning four (the fourth parvovirus found to infect humans)
Ambi: from Latin ambi- meaning on both sides
Brevi: from Latin breve meaning short
Itera: from Latin iterum meaning again
Hepan: from Hepatopancreatic parvovirus, the original name of the founding virus
Penstyl: from Penaeus stylirostris, the host of founding virus
Subfamily demarcation criteria
The two subfamilies, Parvovirinae and Densovirinae, are distinguished primarily by their ability to infect vertebrate versus invertebrate hosts, respectively; a distinction that is generally supported by Bayesian phylogeny of the NS1 protein sequence (Figure 5.Parvoviridae).
In order to be considered for classification in either subfamily, a candidate virus must be judged to represent an authentic parvovirus by meeting certain criteria. Specifically, it must have been isolated and its DNA sequenced or, failing that, it must have been sequenced in tissues, secretions, or excretions of unambiguous host origin, supported by evidence of its distribution in multiple individual hosts in a pattern that is compatible with dissemination by infection. The sequence must be in a single contig, include all nonstructural (NS) and capsid virus protein (VP) coding regions, and reflect the size constraints, motif patterns, and structures typical of the family. This definition does not absolutely require the isolation of a viable virus provided an infectious etiology is supported by the structure and arrangement of the genome, serology, or other biological data. It is designed to allow the inclusion of viruses identified by virus discovery approaches, which typically lack reliable sequences for the telomeric hairpins, while avoiding viral sequence fragments integrated into host genomes or metagenomic data that lack clear host attribution.
Relationships within the family
Members of the two parvovirus subfamilies are distinguished primarily by their vertebrate or invertebrate host range, but this organization is also strongly supported by phylogenetic analysis based on the amino acid sequence of the viral replication initiator protein (Figure 5.Parvoviridae). Outgroups for the analysis included initiator proteins encoded by an anellovirus (torque teno felis virus, AB076003) and a circovirus (chimpanzee faeces asssociated cyclovirus, GQ404850), chosen because they also have linked HuH-family endonuclease and superfamily 3 (SFC) helicase domains and support a linear single-strand displacement replication mechanism that in many ways resembles parvovirus rolling hairpin synthesis. Phylogenetic relationships indicate that the evolutionary roots of viruses in subfamily Densovirinae are extremely ancient when compared to those in subfamily Parvovirinae (Figure 5.Parvoviridae), which is reflected in the greater structural diversity seen among extant densoviruses. Phylogenetic trees also show that an early schism occurred in the evolution of subfamily Densovirinae, creating two ancient branches that lead either to viruses in genera Ambidensovirus and Iteradensovirus or to those in genera Brevidensovirus, Penstyldensovirus and Hepandensovirus (Figures 5.Parvoviridae, 6A.Parvoviridae). In subfamily Parvovirinae more recent branching creates distinct lineage clusters, with one early branch giving rise to the largely heterotelomeric viruses that occupy genera Amdoparvovirus, Protoparvovirus, Aveparvovirus and Bocaparvovirus, while a second branch leads to homotelomeric viruses in genera Copiparvovirus, Dependoparvovirus, Tetraparvovirus and Erythroparvovirus (Figure 6B.Parvoviridae).
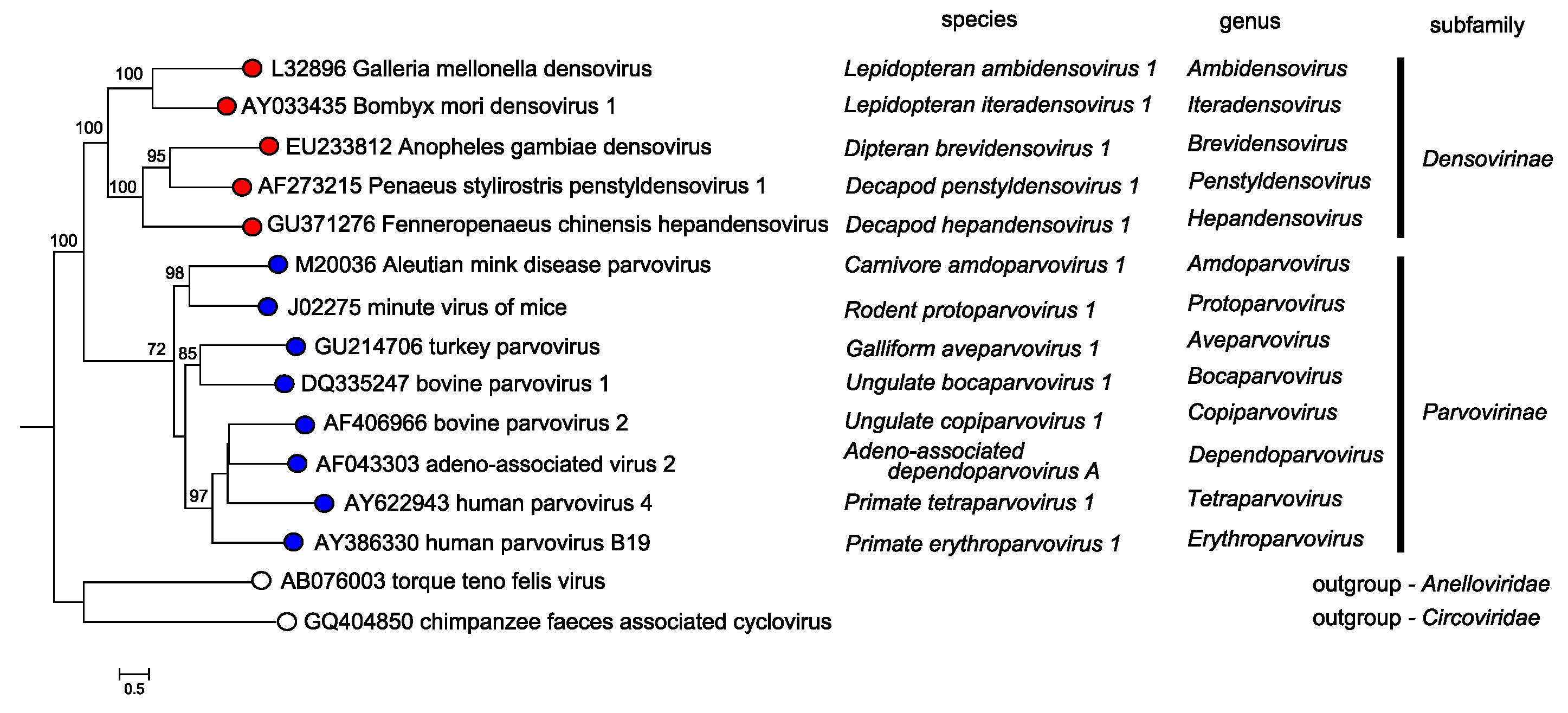 |
| Figure 5. Parvoviridae. Overview of phylogenetic relationships between members of the genera in subfamilies Parvovirinae and Densovirinae. A single exemplar isolate from the type species of each genus is shown. Viruses in subfamily Densovirinae are shown in red, those in subfamily Parvovirinae in blue, and those in outgroups from virus families that use rolling circle replication in yellow. Species names, in italics, follow individual virus names on each coloured branch. Bootstrap support values of 70% or more are indicated at nodes. Trees are based on the amino acid sequences of NS1, which has multiple conserved protein motifs present in all parvoviruses. Taxonomic relationships were determined using the ViCTree pipeline platform (Modha et al., 2018), which automatically selects candidate virus sequences from GenBank. Pairwise distance matrices and multiple sequence alignments were determined using Clustal-omega and a maximum likelihood phylogenetic tree with bootstrap support was generated using RaxML. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
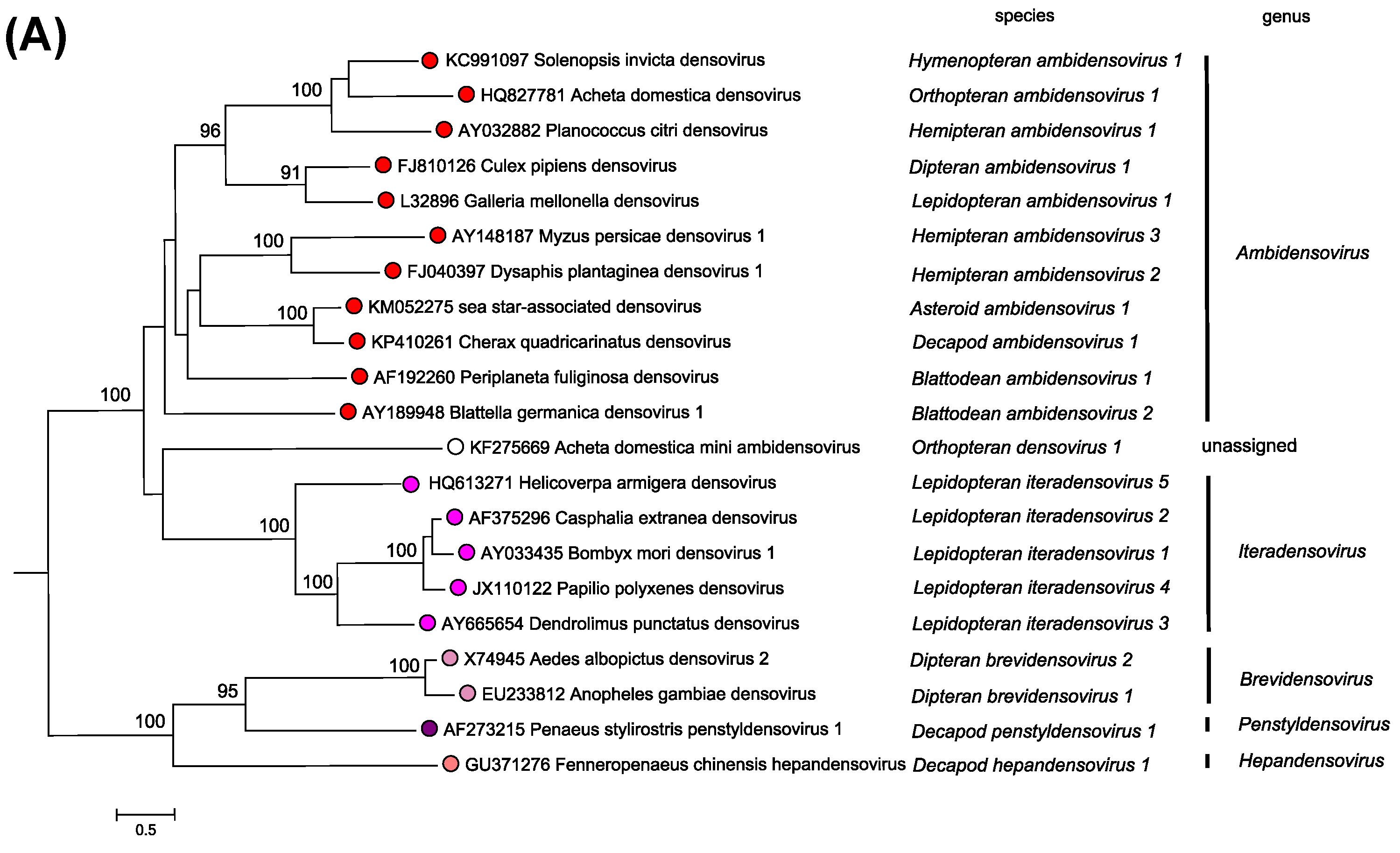 |
| Figure 6A.Parvoviridae. Phylogenetic relationships within the family Parvoviridae. A single exemplar isolate from each species in subfamily Densovirinae is shown in red or red/mauve. Bootstrap support values of 70% or more are indicated at nodes. Phylogeny is based on the amino acid sequences of NS1, as described in the legend to Figure 5.Parvoviridae. This phylogenetic trees and corresponding sequence alignments are available to download from the Resources page. |
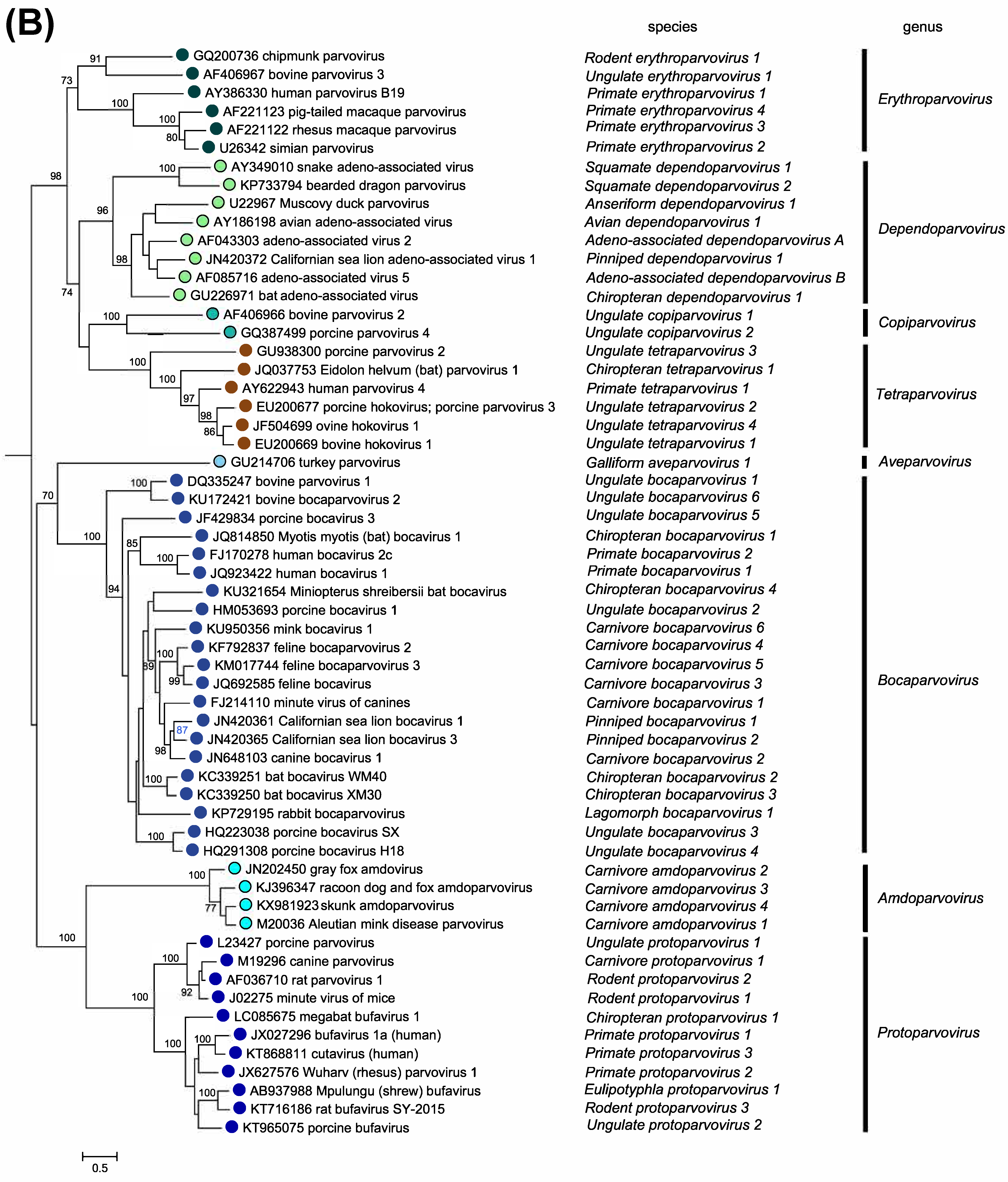 |
| Figure 6B.Parvoviridae. Phylogenetic relationships within the family Parvoviridae. Exemplar isolates from each species in subfamily Parvovirinae are shown in blue or blue/green. Bootstrap support values of 70% or more are indicated at nodes. Phylogeny is based on the amino acid sequences of NS1, as described in the legend to Figure 5.Parvoviridae. This phylogenetic trees and corresponding sequence alignments are available to download from the Resources page. |
Relationships with other taxa
Parvovirus genomes contain just two gene complexes that primarily encode the replication initiator protein, NS1, and a single capsid protein. As discussed previously, NS1 is an HuH endonuclease that cleaves origin structures created at each end of the viral genome using a trans-esterification reaction that introduces a site-specific single strand nick, thus liberating a 3′-nucleotide to prime additional synthesis, but leaving NS1 covalently attached to the new 5′-end of the DNA. This cleavage reaction, the architecture of the nuclease, and its cation-binding and active-site tyrosine protein motifs all closely resemble those of the initiator proteins encoded by other eukaryotic virus families that employ rolling circle replication (RCR) mechanisms (Cotmore and Tattersall 2014, Hickman et al., 2004, Tewary et al., 2013, Tewary et al., 2015). However, the parvovirus molecules are unusual because they lack one of the three evolutionarily conserved HuH nuclease motifs (Koonin and Ilyina 1993), and because they do not catalyze a subsequent trans-esterification reaction that other RCR viruses use to reverse the covalent DNA:protein linkage and join the ends of progeny strands, thus circularizing the displaced DNA. In consequence, parvoviral genomes remain linear and a copy of NS1 is left covalently attached via its active-site tyrosine to the 5′-end of all viral DNA, where it may persist throughout replication, packaging, and virion release (Cotmore and Tattersall 1989).
Previously, some bi-segmented ssDNA viruses that infect silkworms were described as possible parvoviruses (Bombyx mori densoviruses, BmDNV2 and BmDNV3). However, among other differences, these have 2 large genome segments, VD1 (AB033596) and VD2 (S78547), and have now been reclassified in a new family, the Bidnaviridae, and renamed Bombyx mori bidensovirus (BmBDV). It has been suggested that bidnaviruses may have arisen by integration of an ancestral parvovirus genome into a large virus-derived DNA transposon from the polinton family (Krupovic and Koonin 2014).
Up to three iterations of a parvovirus NS1-like sequence have been found in the left-end of aviadenovirus (Adenoviridae) genomes (Kaján et al., 2012), although their significance has yet to be determined.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| porcine parvovirus 7 | KU563733 | PPV7 |
| rat parvovirus 2 | KX272741 | RPV2 |
| Desmodus rotundus parvovirus | KX907333 | Dr-BatPV-1 |
| murine chapparvovirus | MF175078 | MuCPV |
Virus names and virus abbreviations are not official ICTV designations.
To date the above viruses have not been isolated. Their published sequences lack telomere information and have not been validated in any way by cloning and expression. However, these putative virus sequences potentially encode a variant capsid structure and appear to represent founder members of a new genus, provisionally dubbed “genus Chapparvovirus” in the literature. At present we do not know if similar viruses also infect invertebrates, making subfamily assignment uncertain.

