Subfamily: Parvovirinae
Genus: Bocaparvovirus
Distinguishing features
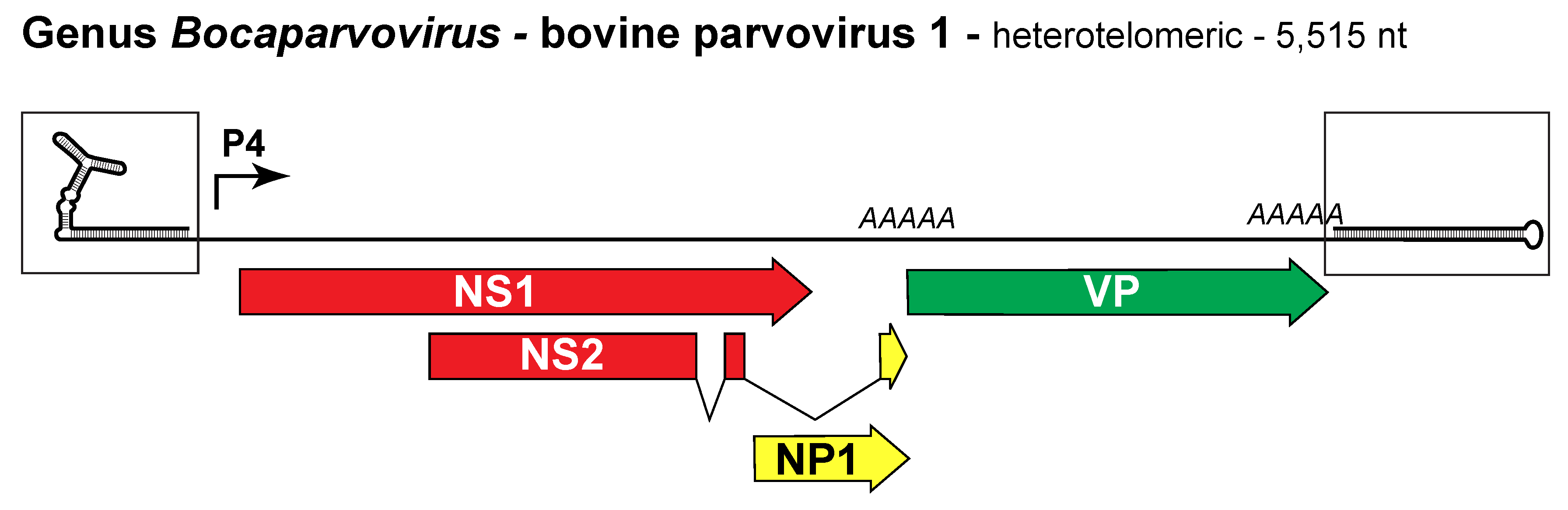
Viruses in this genus are monophyletic and most share >30% NS1 amino acid identity. However, <30% identity values are allowed between certain viruses to accommodate disparities between current and previous analytical methods. Bocaparvoviruses have heterotelomeric genomes of ~5.0–5.5 kb (Figure 1. Bocaparvovirus) (Chen et al., 1986, Sun et al., 2009, Huang et al., 2012). They package predominantly (90–95%) negative-sense DNA and express a unique ~25 kDa ancillary protein called NP1 that is encoded near the middle of the genome, partly overlapping with the C-terminus of NS1 but in a different reading frame. This large genus includes 21 species (Figure 6B. Parvoviridae), members of two of which infect humans. The great majority of these viruses were identified using molecular discovery approaches, and have yet to be isolated.
Virion
See discussion under family description.
In addition to X-ray and cryo-EM image reconstructions of bovine parvovirus 1 (BPV1) (Figures 1. Parvoviridae and 2. Parvoviridae) (Kailasan et al., 2015b), high-resolution cryo-EM reconstructions are available for human bocaviruses 1, 3 and 4 (HBoV1, 3 and 4) (Mietzsch et al., 2017). As for BPV1, these models have density inside the capsid at the base of each 5-fold pore, corresponding to amino acids 29 to 38 from the N-terminus of VP2, which is structurally unique to this genus based on available parvovirus capsid structures.
|
|
|
Figure 1. Bocaparvovirus. Genetic strategy of bovine parvovirus 1 (BPV1), genus Bocaparvovirus. The heterotelomeric genome of BPV1 is shown as a single line terminating in boxed hairpin structures of 148 nt at the left and 146 nt at the right ends of the genome, which are magnified to show potential structure. Segments encoding NS1 are shaded red, those expressed in the capsid proteins are green, and those in the unique bocavirus ancillary protein, NP1, are yellow. A solid arrow (P4) represents the RNA polymerase-II promoter; AAAAA indicates polyadenylation sites. |
Genome organization and replication
Three viruses from this genus, bovine parvovirus 1 (BPV1), human bocavirus 1 (HBoV1), and minute virus of canines (MVC), have been isolated and their DNA cloned into bacterial plasmids to produce infectious clones that have facilitated genetic analysis of their molecular biology. Bocaviruses use a RNA polymerase-II transcriptional promoter near the left end of the genome to generate a single pre-mRNA, which is alternatively spliced, and polyadenylated at two different positions, called (p(A)-proximal) and (p(A)-distal), to generate around 8–14 mature mRNA transcripts (Fasina et al., 2017, Qiu et al., 2007, Chen et al., 2010a). In addition to NS1 and NP1, a series of 1–3 NS ancillary proteins are expressed that contain regions from NS1 and NP1, some of which are required for productive replication (Shen et al., 2015). NP1 is the only known parvoviral ancillary protein that plays a role in RNA processing. It controls splicing of VP-encoding RNAs and read-through of the proximal polyadenylation site, thus controlling expression of a subset of NS ancillary proteins and access to viral capsid sequences (Fasina et al., 2016, Fasina et al., 2017, Zou et al., 2016).
Unlike BPV1 or MVC, the expression strategy of HBoV1 requires splicing to generate full-length forms of NS1. At present HBoV1 appears unique among members of the Parvoviridae in that it expresses a small non-coding RNA polymerase-III transcript (BocaSR) from the 3′-non-coding region of the viral positive-sense strand. This adenovirus VA-like RNA is essential for expression of some viral NS proteins and for viral DNA replication, but unlike adenovirus VA, BocaSR is localized in the nucleus, and does not act through RNA-activated protein kinase R (Wang et al., 2017b).
Biology
BPV1 and MVC were the first two viruses classified in genus Bocaparvovirus. BPV1 causes gastrointestinal and respiratory disease, reproductive failure and conjunctivitis in cattle worldwide. The virus is spread by faecal-oral transmission, and can cross the placenta to infect foetuses, commonly leading to abortion. MVC, also known as canine minute virus (CnMV), is widely distributed in the USA. In adult dogs, infection with MVC is generally sub-clinical, although it can sometimes cause enteritis, but foetal infections commonly lead to neonatal respiratory disease or abortion (Binn et al., 1970).
HBoV1 was identified using a virus discovery approach in respiratory swabs from children with lower respiratory tract infections (Allander et al., 2005). Although it can be the sole causative agent of mild to life-threatening lower respiratory tract infections, especially in young children (<5 years of age), it is, due to virus persistence, often also co-detected with other viruses causing subsequent respiratory infections (Qiu et al., 2017, Jartti et al., 2012, Moesker et al., 2015). It infects >80% of humans, generally early in life, and is present in most regions of the world. In tissue culture, HBoV1 is only able to productively infect terminally differentiated, polarized human airway epithelial cells that are cultured at an air-liquid interface (Huang et al., 2012, Dijkman et al., 2009). It is, therefore, unlike most other parvoviruses because it can replicate in non-dividing cells, which it achieves using host synthetic machinery made available in response to a DNA damage response that the virus evokes (Deng et al., 2016). By 2010, three more HBoV1-like viruses, named HBoV2c–4, had been identified in human stool samples from children with gastrointestinal illness (Kapoor et al., 2009, Arthur et al., 2009, Kapoor et al., 2010). There are inconsistent data linking HBoV2c with gastroenteritis, and little evidence that the less abundant HBoV3 and 4 forms are pathogenic (Paloniemi et al., 2014, Mori et al., 2013).
Most of the other known bocaviruses were identified in faecal and/or respiratory samples by virus discovery approaches, although some, less commonly, have also been detected in serum and tissues. Epidemiological studies indicate that many are widespread in their respective host populations, but are rarely pathogenic.
Species demarcation criteria
Viruses within a species are monophyletic and encode replication initiator proteins (called NS1 or Rep1, 68, or 78) that show >85% amino acid sequence identity.
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
dromedary camel bocaparvovirus 1 |
DBoV1 |
|
|
dromedary camel bocaparvovirus 2 |
DBoV2 |
|
|
rat bocavirus |
RBoV |
|
|
murine bocavirus |
MuBoV |