Subfamily: Parvovirinae
Genus: Tetraparvovirus
Distinguishing features
Genus Tetraparvovirus was established in 2014 to accommodate a cluster of monophyletic viruses identified using virus discovery approaches. The genus is part of a major clade in subfamily Parvovirinae that includes members of genera Dependoparvovirus, Erythroparvovirus and Copiparvovirus (Figure 6B. Parvoviridae). Tetraparvovirus genomes are around 5 kb, but no telomere sequence information is available and none of the viruses have been isolated or propagated in culture. The founder virus, human parvovirus 4 (PARV4), was identified in 2005 in nuclease-digested plasma from patients with acute viral infection syndrome (Jones et al., 2005) and has been detected, with widely varying frequencies, in plasma samples worldwide. Its clinical associations remain uncertain. There are 6 species in the genus, including viruses that infect primates, bats or ungulates (pigs, sheep or cattle).
Virion
See discussion under family description.
Genome organization and replication
See discussion under family description. PARV4 is the exemplar virus of the type species, Primate tetraparvovirus 1. This species includes three distinct PARV4 genotypes (G1 AY622943, G2 DQ873390, and G3 EU874248) that have <3% amino acid sequence divergence, plus a virus that infects chimpanzees (HQ113143). Current knowledge of PARV4 has been reviewed in detail elsewhere (Qiu et al., 2017, Matthews et al., 2017). G1 is the predominant form circulating in Europe and North America at the moment, but G2 (sometimes known as PARV5) also circulates in these areas and is most common in those who were likely infected in the 1980s, suggesting that it may have been the predominant form at that time. G2 and G2-like forms also circulate in Asia and Brazil, whereas G3 is the major strain found in Africa.
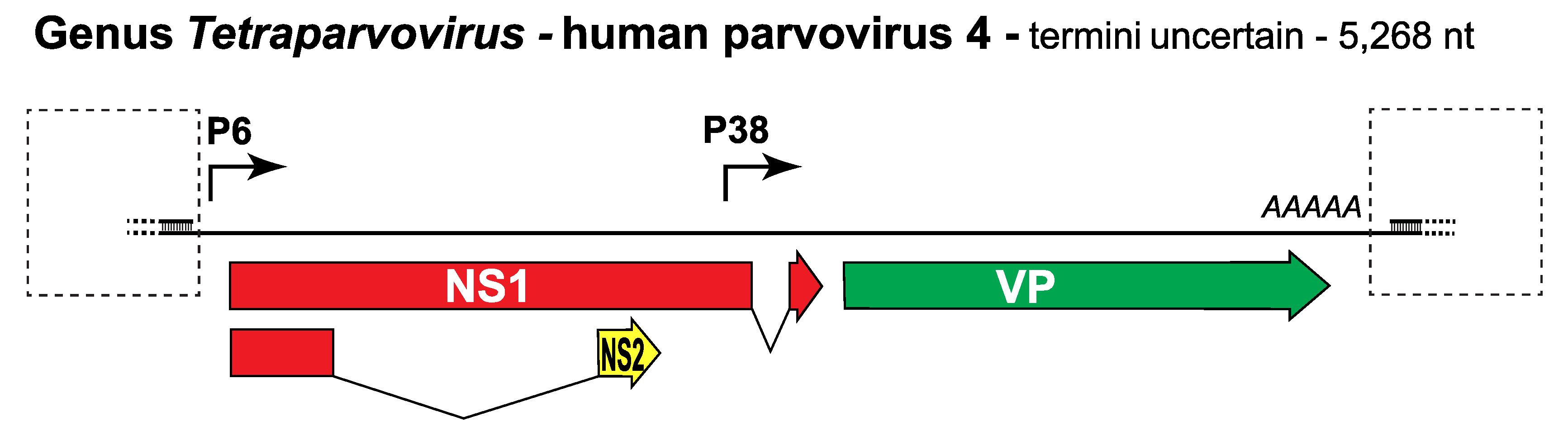
Since no complete genomes are available, the PARV4 gene expression profile has been explored provisionally by transfecting its coding sequences into tissue culture cells. Because DNA replication commonly affects parvovirus transcription patterns, these sequences were first inserted between adeno-associated virus 5 (AAV5) terminal hairpins and the resulting hybrid induced to replicate by co-transfection into 293 cells with constructs encoding AAV5Rep78 and adenovirus helper factors (Lou et al., 2012). Two promoters, P6 and P38, were identified, which generate transcripts encoding NS and VP proteins respectively (Figure 1. Tetraparvovirus). Some P6 transcripts predicted to initiate in the same frame as NS1 are then spliced into an alternate frame, potentially giving rise to a small NS2 protein, although this product was not detected. Messenger RNAs encoding VP2 and NS1 were observed, generating proteins with molecular masses of ~80 and ~65 kDa respectively, but it remains uncertain how the predicted VP1 coding sequences, which include a VP1 specific region of at least 261 amino acids containing a putative PLA2 domain, are accessed.
|
|
|
Figure 1. Tetraparvovirus. Genetic strategy of human parvovirus 4 (PARV4), genus Tetraparvovirus. The coding sequence of PARV4 is shown as a single line with dashed boxes indicating the positions of predicted terminal hairpin structures. Open reading frames expressed in NS1 are colored red, those encoding the capsid proteins are green, and one that is unique to the NS2 ancillary protein is yellow. Solid arrows indicate transcriptional promoters; AAAAA indicates the single polyadenylation site. |
Biology
See discussion under family description. Little is known about the biology or clinical significance of any members of this genus. PARV4 genomes have been detected in plasma during acute infection but often with low viral loads (<3 × 104 genome copies/ml) and viremia appears protracted, typically lasting from one to several months. Viral genomes have also been detected in some liver and bone marrow samples, although where the virus replicates remains unknown. PARV4 DNA-positive or PARV4 IgG-positive plasmas are rare in the general population of North America and Europe, but occur more frequently in individuals carrying other blood-borne viruses, most notably human immunodeficiency virus, hepatitis B virus or hepatitis C virus, or who have behavioural risk factors for parenteral infection such as intravenous drug use or reliance on hemoderivatives from pooled human plasma (Lahtinen et al., 2011, Sharp et al., 2009). However, in Africa PARV4 is endemic and its epidemiology appears very different, with around 30–50% of the general population in sub-Saharan and South Africa testing seropositive for PARV4 IgG. Viremia is also detected frequently, for example in one study 8.6% of young asymptomatic children in Ghana were DNA-positive (Panning et al., 2010), and viral DNA has been found in nasal and stool samples from African children, suggesting that in this locale transmission is likely by foodborne, respiratory, or contact-mediated routes.
PARV4 genotypes 1 and 2 predominate in Europe and North Africa, whereas G3 is the major form in Africa; thus genetic differences that affect virus biology may contribute to these extreme epidemiological disparities. Alternatively, characteristics of the host population may be critical. Accordingly, much current research is directed at clarifying virus susceptibility and transmission routes. For example, a recent study from Brazil, where the circulating G2 virus might be predicted to follow a parenteral route/high-risk group pattern, revealed that around 6% of individuals infected with human T-cell lymphotropic virus (HTLV) were also positive for PARV4 DNA, but the majority of these co-infected individuals had no history suggesting a parenteral transmission route, indicating that additional factors or routes are likely involved in this locale (Slavov et al., 2017).
Tetraparvoviruses that infect non-human hosts also appear endemic. For example, 63% of chimpanzees and 18% of gorillas from a group of 73 wild-caught apes sampled in Cameroon tested seropositive for antibodies against the chimpanzee virus in species Primate tetraparvovirus 1 (Beierwaltes 1991), while viruses in the 4 ungulate species, Ungulate tetraparvovirus 1, Ungulate tetraparvovirus 2, Ungulate tetraparvovirus 3, and Ungulate tetraparvovirus 4, were commonly detected in the serum and tissues of livestock (Lau et al., 2008, Tse et al., 2011). Similarly, a bat virus (in species Chiropteran tetraparvovirus 1) was detected at high concentration in blood samples and tissues from Eidolon helvum bats in Ghana. This virus was particularly abundant in samples from spleen and kidney, suggesting these organs as likely sites for viral replication (Canuti et al., 2011).
Species demarcation criteria
Viruses within a species are monophyletic and encode replication initiator proteins (called NS1 or Rep1, 68, or 78) that show >85% amino acid sequence identity.