Family: Parvoviridae
Subfamily: Densovirinae
Distinguishing features
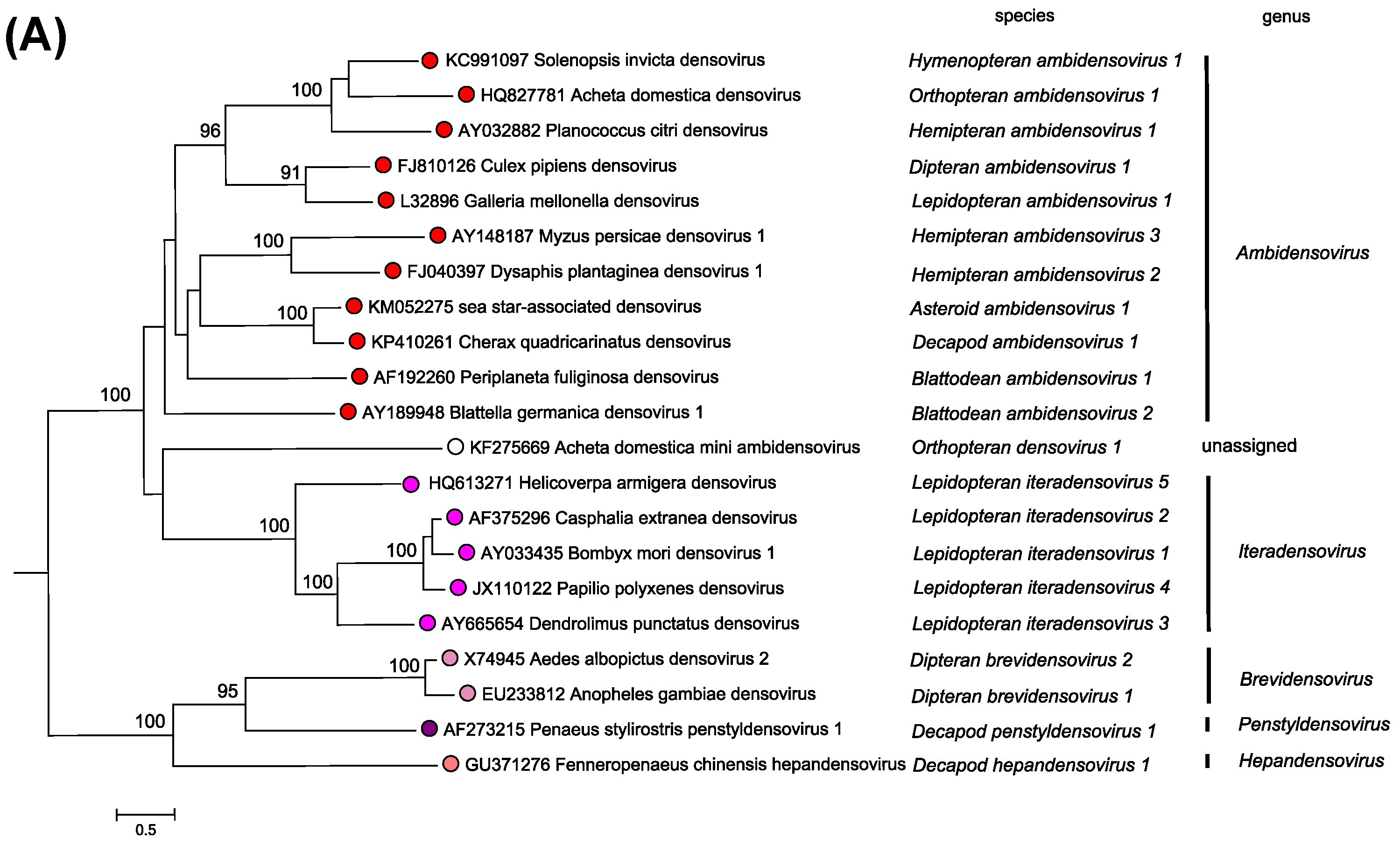
Viruses in this subfamily infect invertebrate hosts (currently known to infect insects from 6 orders, Blattodea, Diptera, Hemiptera, Hymenoptera, Lepidoptera, and Orthoptera), decapod crustaceans (shrimp and crayfish), and echinoderms (sea stars). Most of the known viruses are highly pathogenic, and infected tissues often display distinctive nuclear hypertrophy, caused by the accumulation of large virion masses, and cytoplasmic paracrystalline virion arrays. The structures and phylogeny of known viruses appear more diverse than their counterparts in subfamily Parvovirinae (Figures 1. Parvoviridae, 2. Parvoviridae, 3. Parvoviridae, 5. Parvoviridae and 6. Parvoviridae). Currently there are 5 recognized genera and one unassigned species (Orthopteran densovirus 1) (Figure 1. Densovirinae), which segregate into two lineages. One lineage includes members of the Ambidensovirus and Iteradensovirus genera and the unassigned species Orthopteran densovirus 1, which all have a PLA2 domain in their VP1 proteins and possess complex viral capsids with up to four VPs. Until recently members of this lineage were only known to infect insects. The second lineage encompasses members of genus Brevidensovirus, which infect mosquitos, and viruses from the Hepandensovirus and Penstyldensovirus genera that appear restricted to decapod hosts from the family Penaeidae. Despite their diverse capsid proteins, no members of this branch have been demonstrated to encode a PLA2 domain.
Genus demarcation criteria
All viruses in a genus should be monophyletic and encode NS1 proteins that are >30% identical to each other at the amino acid sequence level. However, at present full coding sequences are only available for a small number of economically significant densoviruses, while there are over a million potential invertebrate host species, so that existing knowledge may drastically understate the diverse nature of this subfamily. To prevent excessive fragmentation of the current phylogenetic tree, in some genera the >30% identity requirement is relaxed to allow inclusion in the genera of monophyletic viruses with conspicuously similar characteristics, sometimes from host orders separated by large evolutionary distances.
|
Figure 1. Densovirinae. Phylogenetic relationships within the subfamily Densovirinae. The phylogeny is based on the amino acid sequences of NS1 from a single exemplar isolate from each species in the subfamily, as described in the legend to Figure 5. Parvoviridae. Bootstrap support values of 70% or more are indicated at nodes. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |