Family: Pospiviroidae
Francesco Di Serio, Robert A. Owens, Shi-Fang Li, Jaroslav Matoušek, Vicente Pallás, John W. Randles, Teruo Sano, Jacobus Th. J. Verhoeven, Georgios Vidalakis, and Ricardo Flores
The citation for this ICTV Report chapter is the summary published as Di Serio et al. (2021):
ICTV Virus Taxonomy Profile: Pospiviroidae, Journal of General Virology, 102 (2): 001543
Corresponding author: Francesco Di Serio (francesco.diserio@ipsp.cnr.it)
Edited by: F. Murilo Zerbini and Sead Sabanadzovic
Posted: November 2020
PDF: ICTV_Pospiviroidae.pdf
Summary
Members of the family Pospiviroidae have a genome composed of a single-stranded circular RNA molecule that adopts a rod-like or quasi rod-like conformation containing a central conserved region (CCR) involved in their replication. These viroids lack the hammerhead ribozymes that are typical of members of the family Avsunviroidae. The family Pospiviroidae includes five genera: Apscaviroid, Cocadviroid, Coleviroid, Hostuviroid and Pospiviroid (Table 1.Pospiviroidae).
Table 1.Pospiviroidae. Characteristics of members of the family Pospiviroidae
| Characteristic | Description |
| Typical member | potato spindle tuber viroid (V01465), species Pospiviroid fusituberis |
| Genome | Single-stranded, circular RNA of 246–375 nt that adopt |
| Replication | Mediated by nuclear DNA-dependent RNA polymerase II, redirected to recognize RNA templates, through an asymmetric RNA-RNA rolling-circle mechanism in which only oligomeric RNAs of (+) polarity are cleaved by a type-III RNase and circularized by DNA ligase 1 redirected to act on RNA substrates |
| Translation | Absent |
| Host range | Plants (dicots and some monocots) |
| Taxonomy | Five genera, collectively including 40 species |
Physicochemical and physical properties
Members of family Pospiviroidae have genomes composed of circular single-stranded RNAs ranging in size from about 246 to 375 nt. The G+C content is >50%. These circular RNAs may assume rod-like or quasi-rod-like conformations in silico or in vitro (Giguère et al., 2014, Steger and Perreault 2016) (Figure 1.Pospiviroidae), with both indirect (Wassenegger et al., 1994, Haseloff et al., 1982, Semancik et al., 1994, Fadda et al., 2003) and direct evidence supporting a similar conformation for potato spindle tuber viroid (PSTVd) in vivo (López-Carrasco and Flores 2017). Members of the family Pospiviroidae are soluble in 2 M LiCl (Navarro and Flores 1997).
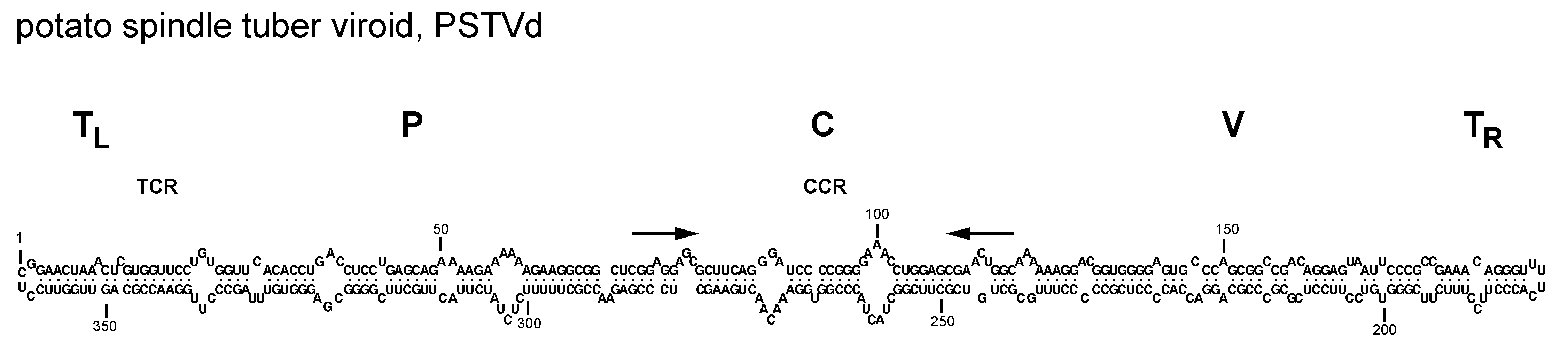 |
| Figure 1.Pospiviroidae. Primary and proposed rod-like secondary structure of minimum free energy for the reference sequence variant of potato spindle tuber viroid (PSTVd, NC_002030). The secondary structure has been calculated using the RNAfold server available in the ViennaRNA Web Services (http://rna.tbi.univie.ac.at/) |
Nucleic acid
RNAs of both polarities are present in the infected hosts, with the strand accumulating at higher level in vivo arbitrarily considered to be of (+) polarity. Circular forms are exclusively of (+) polarity. In contrast, the RNAs of (-) polarity are exclusively linear and accumulate predominantly as oligomeric molecules that serve as replication intermediates (Branch and Robertson 1984, Flores et al., 2017). The compact secondary structure adopted by the circular RNAs likely increases their resistance to degradation by host RNases. Alternatively, the circular RNA may also form metastable structures (Henco et al., 1979, Riesner et al., 1979) with hairpins that are functionally important (Riesner 1991). Prominent among these is hairpin I (HPI) formed by the upper strand of the central conserved region (CCR) and the flanking inverted repeats (Riesner et al., 1979, Visvader and Symons 1985, Polivka et al., 1996). Despite the sequence variability among viroids of different genera, HPI is conserved in all members of the family Pospiviroidae and is most likely involved in their replication (Figure 2.Pospiviroidae). Elements of tertiary structure, including the loop E motif previously described in 5S RNA, have been identified in PSTVd, with proposed roles in viroid replication, accumulation, or movement (Gas et al., 2008, Wang et al., 2018, Wu et al., 2019).
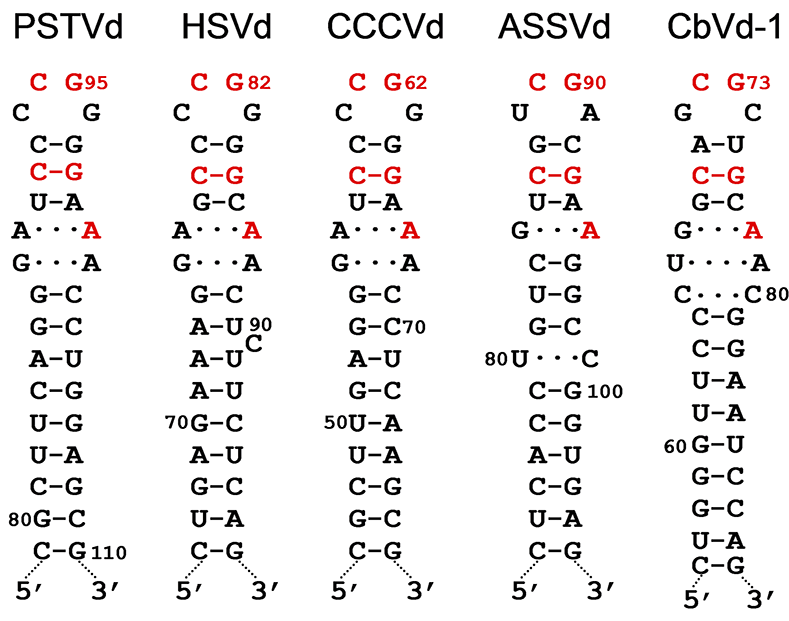 |
| Figure 2.Pospiviroidae. Hairpin I structures of typical members of species of five genera in the family Pospiviroidae. PSTVd, potato spindle tuber viroid; HSVd, hop stunt viroid; CCCVd, coconut cadang-cadang viroid; ASSVd, apple scar skin viroid, and CbVd-1, Coleus blumei viroid 1. Red fonts indicate conserved nucleotides in structurally similar positions. Continuous and broken lines represent Watson-Crick and non-canonical base pairs, respectively. Adapted with modifications from (Gas et al., 2007). |
Genome organization and replication
Viroids in this family share five structural/functional domains within the proposed rod-like secondary structure of minimal free energy adopted by the circular genomic RNA: C (central), P (pathogenic), V (variable) and TL and TR (terminal left and right, respectively) (Figure 3.Pospiviroidae) (Keese and Symons 1985). The C domain contains a central conserved region (CCR) formed by two sets of conserved nucleotides located in the upper and lower strands, with those of the upper strand being flanked by an imperfect inverted repeat that interact to form the HPI (Figure 2.Pospiviroidae) or, in oligomers, a double-stranded structure possibly involved in replication (Gas et al., 2007), although an alternative mechanism has been proposed (Baumstark et al., 1997). Two other conserved sequence elements are the terminal conserved region (TCR), found in all members of the genera Pospiviroid and Apscaviroid as well as the two largest members of the genus Coleviroid, and a terminal conserved hairpin (TCH) present in all members of the genera Hostuviroid and Cocadviroid (Figure 3.Pospiviroidae) (Koltunow and Rezaian 1989, Puchta et al., 1988, Flores et al., 1997). The conservation of these two elements in both sequence and location in the left terminal domain suggests their involvement in critical functions. Together with CCR, TCR and TCH are used for taxonomic purposes (Di Serio et al., 2014). Conservation of structural elements among different viroids has been also observed in solution by the selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) and their relevance for classification purposes confirmed (Giguère and Perreault 2017). Coconut cadang-cadang viroid (CCCVd) is unusual in that it occurs as RNAs of different sizes, the larger ones containing repetitions of a portion of the sequence of the smallest one (Haseloff et al., 1982); a similar situation has been found in citrus exocortis viroid (CEVd) (Semancik et al., 1994, Fadda et al., 2003). Some viroids, the most striking examples being columnea latent viroid (CLVd) and Australian grapevine viroid (AGVd), appear to have arisen by intermolecular RNA recombination since they seem to consist of a more or less complex mosaic of sequences present in other viroids (Rezaian 1990, Hammond et al., 1989). Direct evidence in support of an ongoing recombination process has come from studies on the dynamic distribution over time of coleus viroids co-infecting individual plants (Spieker 1996).
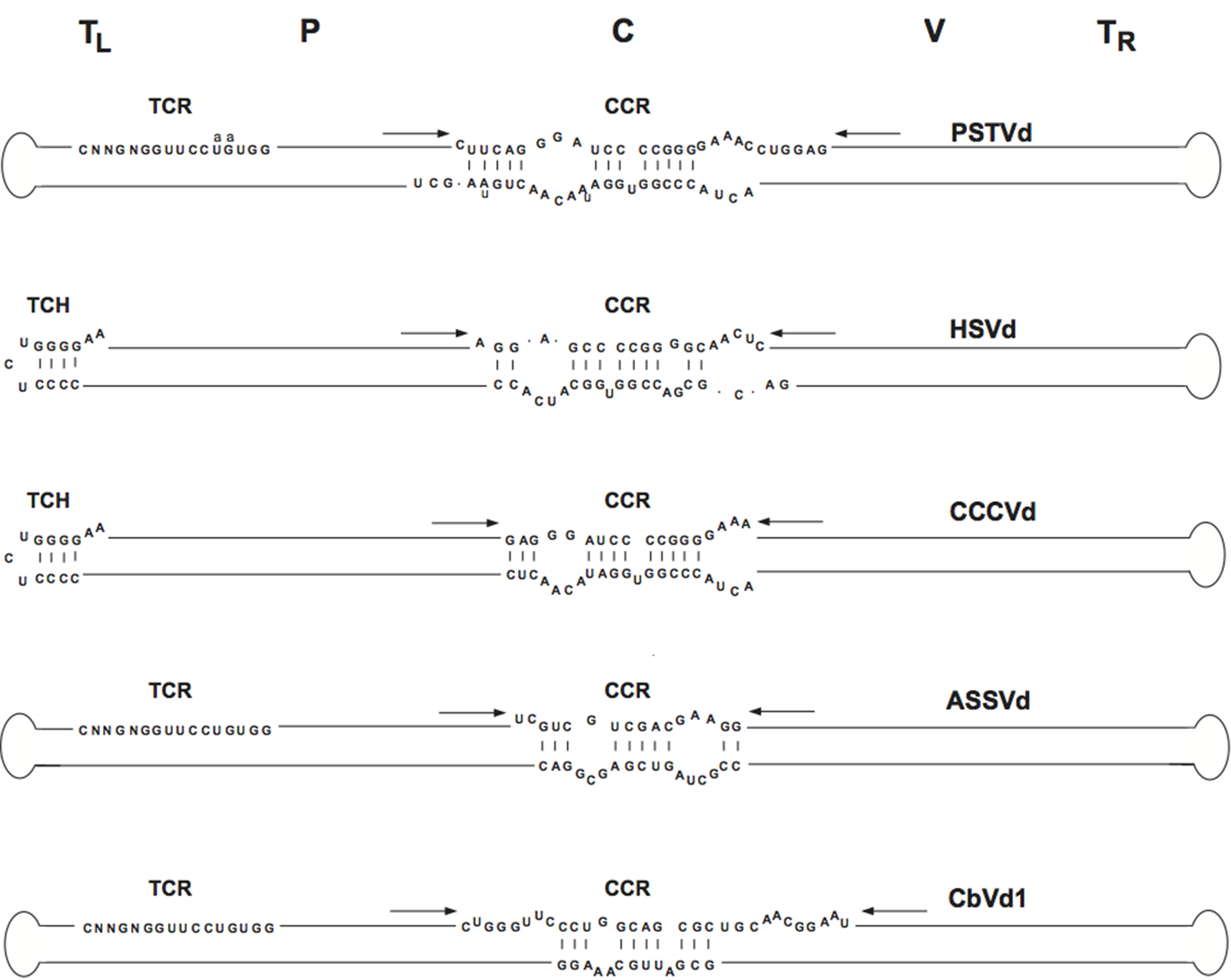 |
| Figure 3.Pospiviroidae. Rod-like structure models for viroids in the different genera of the family Pospiviroidae. The abbreviations of potato spindle tuber viroid (PSTVd), hop stunt viroid (HSVd), coconut cadand-cadang viroid (CCCVd), apple scar skin viroid (ASSVd), and Coleus blumei viroid 1 (CbVd-1), belonging to type species of each genus, are indicated on the right, and the approximate location of the five domains, C (central), P (pathogenic), V (variable) and TL and TR (terminal left and right), at the top of the figure. The core nucleotides (nt) of the central conserved region (CCR), terminal conserved region (TCR) and terminal conserved hairpin (TCH) are shown. Arrows indicate flanking sequences which form, together with the core nt of the CCR upper strand, imperfect inverted repeats. The core nt of the HSVd CCR have been defined by comparison with columnea latent viroid, which on the basis of other criteria (presence of the TCR, absence of the TCH and overall sequence similarity) is included in the genus Pospiviroid. Substitutions found in the CCR and TCR of iresine viroid 1, a member of the genus Pospiviroid, are indicated in lowercase. In the genus Coleviroid, the TCR only exists in the two largest members Coleus blumei viroid 2 and Coleus blumei viroid 3. Adapted with modifications from (Flores et al., 1997). |
Replication studies have been done mostly with PSTVd, and the inferred mechanism (see below, Figure 1.Viroids) is presumed to operate in the other members of the family as well (Flores et al., 2017). Based on the characterization of viroid RNAs extracted from infected tissues, an asymmetric rolling-circle mechanism based on RNA intermediates has been proposed for the replication of pospiviroids (Branch and Robertson 1984, Grill and Semancik 1978, Ishikawa et al., 1984, Feldstein et al., 1998, Daròs and Flores 2004), which takes place in the nucleus (Spiesmacher et al., 1983, Qi and Ding 2003a). Oligomeric viroid RNAs of both polarities are synthesized by RNA polymerase II (Mühlbach and Sänger 1979, Rackwitz et al., 1981, Flores and Semancik 1982, Schindler and Mühlbach 1992, Warrilow and Symons 1999, Bojić et al., 2012) that is conscripted to transcribe RNA templates instead of its physiological DNA templates. Transcription likely starts at a specific site, with that for the PSTVd (-) strand proposed to be located in the left terminal loop of the rod-like (+) strand (Kolonko et al., 2006). No information is available on the initiation site for synthesis of the (+) strands of PSTVd and other pospiviroids. Oligomeric (+) RNAs are cleaved at specific site(s) by an RNase of group III, generating monomers that are circularized by DNA ligase 1 (Nohales et al., 2012, Flores et al., 2014), the second normally DNA-dependent host enzyme forced to use RNA as substrate.
Biology
Host range
Members of the family Pospiviroidae naturally infect either a limited number or a wide range of angiosperm plant species, mostly dicotyledons. The natural hosts of CCCVd and coconut tinangaja viroid (CTiVd) are monocotyledons (Škorić 2017).
Symptoms
Some infections have devastating effects; for example, CCCVd has killed millions of coconut palms in the Philippines (Randles 2003). Other infections result in epinasty, rugosity, chlorosis and necrosis on leaves, shortening of stem internodes leading to stunted phenotypes, bark scaling, cracking or gumming, deformation and color alterations of fruits and storage organs, and delays in foliation, flowering and ripening. A few viroids induce only mild or no symptoms at all. Moreover, symptoms are strongly influenced by the viroid-host combination, with several members of the family inducing severe symptoms in one crop (i.e. tomato) and remaining symptomless in others (i.e. ornamental species) (Verhoeven et al., 2017). Symptom expression is generally favored by high temperature and light intensities. Molecular determinants of specific symptoms have been mapped in the genome of members of family Pospiviroidae, such as PSTVd (Schnölzer et al., 1985, Sano et al., 1992, Qi and Ding 2003b) and hop stunt viroid (HSVd) (Reanwarakorn and Semancik 1998, Palacio-Bielsa et al., 2004).
Movement
After entry into a host cell, members of the family Pospiviroidae must be transported into the nucleus, where they replicate and accumulate. PSTVd movement into this organelle appears directed by signal(s) contained in the viroid RNA (Zhao et al., 2001) through a cytoskeleton-independent process that is mediated by a specific and saturable receptor and involves recognition of a conserved sequence and/or structural motif in the upper CCR (Woo et al., 1999). Mutational analysis of PSTVd has shown that many of the loops in its rod-like secondary structure (Figure 1.Pospiviroidae) play a role in movement from cell-to-cell and across tissue boundaries, possibly by creating pockets that bind host proteins (Ding et al., 1997, Qi et al., 2004). PSTVd moves systemically through the phloem (Palukaitis 1987, Zhu et al., 2001), a process likely driven by plant proteins. In vitro, HSVd can form a ribonucleoprotein complex with phloem protein 2, a dimeric lectin able to move from cell-to-cell through plasmodesmata and toward sink tissues in the assimilate stream (Gómez and Pallás 2001, Owens et al., 2001, Gómez and Pallás 2004). These properties, together with its ability to bind RNA, suggest that phloem protein 2 facilitates the systemic movement of viroids. Access of PSTVd to floral and vegetative meristems is limited (Zhu et al., 2001), most likely by RNA silencing (Di Serio et al., 2010). The viroid can overcome this defense strategy during specific developmental stage(s), thus making possible transmission through seeds in tomato (Matsushita et al., 2011). The ability of chrysanthemum stunt viroid (CSVd) to invade the shoot apical meristem (SAM) is dependent on the host and the temperature (Matsushita and Shima 2015, Zhang et al., 2015)
Transmission
All members of the family Pospiviroidae are transmitted by vegetative propagation of infected plants. In addition, many can be transmitted mechanically. Some members are also transmitted by seed and pollen. With the exclusion of tomato planta macho viroid (TPMVd), aphid transmission seems negligible. Transmission of some member of the family by bumblebees and whiteflies has been reported, but the relevance of these transmission pathways in nature remains unclear (Škorić 2017, Walia et al., 2015).
Cross-protection and synergism
Cross-protection has been reported between closely related pospiviroids (Niblett et al., 1978) as well as between mild and severe strains of PSTVd (Khoury et al., 1988). RNA silencing has been proposed to mediate this phenomenon. Pronounced developmental alterations have been observed in citrus plants co-infected by citrus viroid V (CVd-V) and either citrus bent leaf viroid (CBLVd) or citrus dwarfing viroid (CDVd), two members of the genus Apscaviroid (Serra et al., 2008). Synergistic and antagonistic effects between different viroids infecting citrus have also been observed in long-term field assays (Semancik et al., 1991, Vernière et al., 2006), but the molecular mechanisms underlying these interactions are still unknown.
Derivation of names
Apscaviroid: from the type species, Apple scar skin viroid
Cocadviroid: from the type species, Coconut cadang-cadang viroid
Coleviroid: from the type species Coleus blumei viroid
Hostuviroid: from the type species, Hop stunt viroid
Pospiviroidae, Pospiviroid: from the type species, Potato spindle tuber viroid (genus Pospiviroid).
Genus demarcation criteria
The type of CCR and the presence/absence of a TCR or TCH in the rod-like secondary structure of the (+) polarity strand (Figure 3.Pospiviroidae) are the primary criteria used to distribute the various species among five genera (Pospiviroid, Hostuviroid, Cocadviroid, Apscaviroid and Coleviroid). There are basically three types of CCRs exemplified by those of PSTVd, apple scar skin viroid (ASSVd) and Coleus blumei viroid 1 (CbVd-1). Species in genera Pospiviroid, Hostuviroid and Cocadviroid share an identical subset of nucleotides within their CCRs and are, therefore, more closely related to one another than to members of the genera Apscaviroid and Coleviroid (Figure 2.Pospiviroidae). A TCR and TCH have never been found together in the same viroid.
Relationships within the family
Members of the family Pospiviroidae are phylogenetically related to each other, and the clustering observed in phylogenetic trees inferred from their complete sequences is consistent with the classification into genera according to the genus demarcation criteria described above (Figure 4.Pospiviroidae).
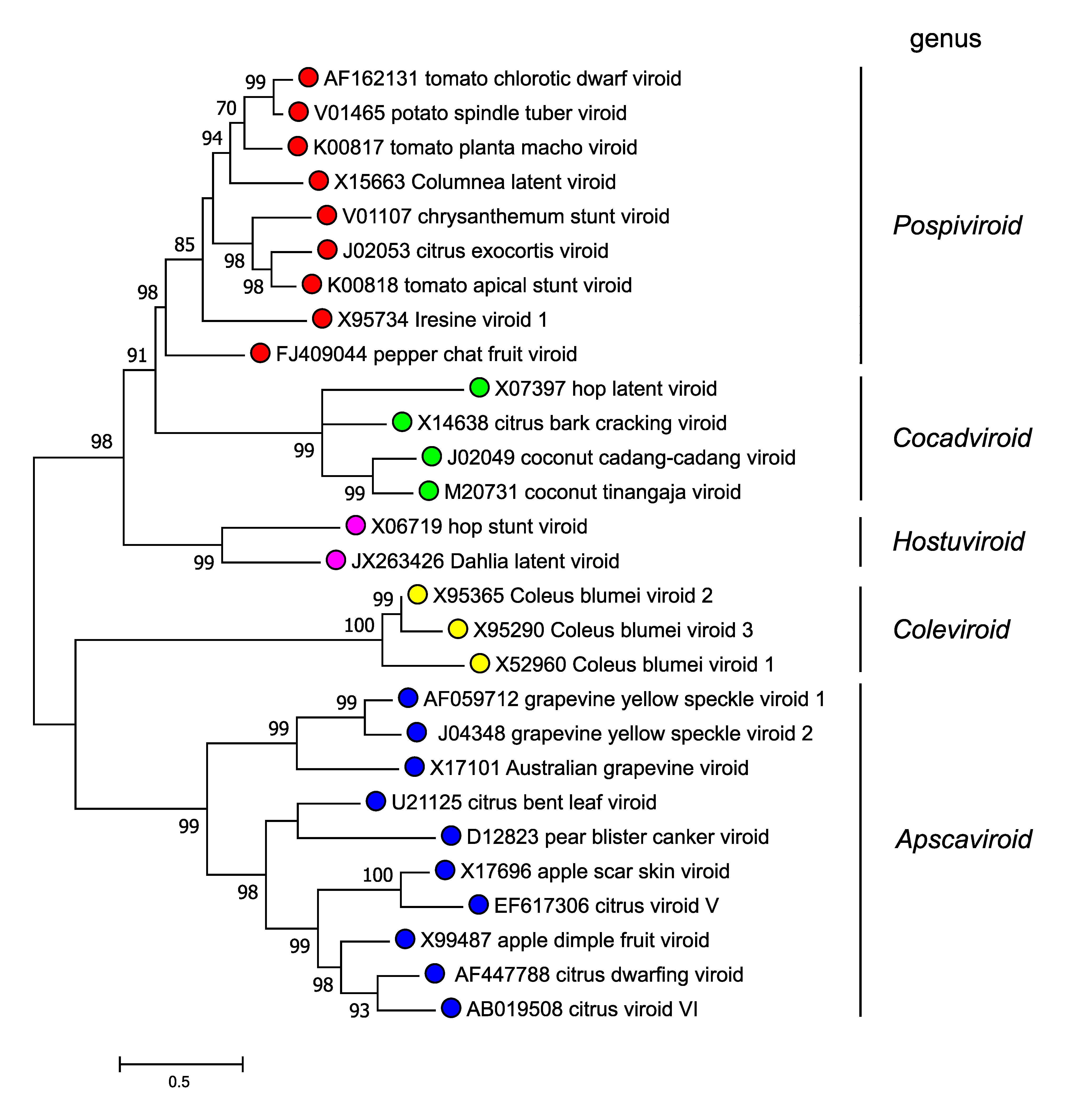 |
| Figure 4.Pospiviroidae. Maximum likelihood phylogenetic tree inferred with the reference variants of the species currently classified in the five genera of the family Pospiviroidae. The alignment was performed with the ClustalW program (gap opening and gap extension penalties 15 and 6.66, respectively). Bootstrap values >70% (generated by 1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). (Navarro, Chiumenti, Flores and Di Serio, unpublished). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the family Pospiviroidae share their small size, circularity and rod-like secondary structure of the genomic RNAs with members of the family Avsunviroidae and with certain viroid-like satellite RNAs associated with viruses. However, the latter two groups of infectious RNAs contain ribozymes, which are absent in members of the family Pospiviroidae, and do not contain a CCR, TCR or TCH.

