Family: Avsunviroidae
Francesco Di Serio, Shi-Fang Li, Jaroslav Matoušek, Robert A. Owens, Vicente Pallás, John W. Randles, Teruo Sano, Jacobus Th. J. Verhoeven, Georgios Vidalakis and Ricardo Flores
The citation for this ICTV Report chapter is the summary published as Serio et al., (2018):
ICTV Virus Taxonomy Profile: Avsunviroidae, Journal of General Virology, 99: 611–612.
Corresponding author: Francesco Di Serio (francesco.diserio@ipsp.cnr.it)
Edited by: F. Murilo Zerbini and Sead Sabanadzovic
Posted: May 2018, updated June 2020
PDF: ICTV_Avsunviroidae.pdf
Summary
Members of the family Avsunviroidae have a genome composed of a single-stranded, circular RNA that adopts a rod-like or a branched conformation and can form, in the strands of either polarity, hammerhead ribozymes involved in their replication in plastids through a symmetric RNA-RNA rolling-circle mechanism (Table 1. Avsunviroidae). These viroids lack the central conserved region typical of members of the family Pospiviroidae. The family Avsunviroidae includes three genera, Avsunviroid, Pelamoviroid and Elaviroid, with a total of five species.
Table 1. Avsunviroidae. Characteristics of members of the family Avsunviroidae.
| Characteristic | Description |
| Typical member | avocado sunblotch viroid (J02020), species Avsunviroid albamaculaperseae |
| Genome | Single-stranded, circular RNA of 246–434 nt that can form hammerhead ribozymes in the strands of either polarity. The (+) polarity is arbitrarily assigned to the most abundant strand in vivo |
| Replication | Mediated by a nuclear-encoded plastid RNA polymerase, which generates complementary (-) oligomeric viroid RNAs through a symmetric rolling-circle mechanism. These replicative intermediates are co-transcriptionally self-cleaved by the embedded hammerhead ribozymes, producing monomeric (-) strands that are circularized by a tRNA ligase. The second part of the mechanism is symmetric with respect to the first. |
| Host range | Plants (dicots) |
| Taxonomy | Three genera, collectively including five species |
Physicochemical and physical properties
Members of family Avsunviroidae are composed of circular single-stranded RNAs ranging in size from 246 to 434 nt. The G+C content is >50% with the exception of avocado sunblotch viroid (ASBVd) with only 38% G+C. Circular RNAs may assume rod-like, quasi-rod-like or branched conformations in silico or in vitro (Symons 1981, Hernández and Flores 1992, Bussière et al., 2000, Fadda et al., 2003, Zhang et al., 2014) (Figure 1. .Avsunviroidae), with indirect (Fadda et al., 2003, Ambrós et al., 1998, de la Peña et al., 1999, López-Carrasco et al., 2016, Rakowski and Symons 1989) or direct (López-Carrasco and Flores 2017) evidence supporting similar conformations in vivo. The plus (+) strands of peach latent mosaic viroid (PLMVd), chrysanthemum chlorotic mottle viroid (CChMVd), and apple hammerhead viroid (AHVd) adopt multibranched conformations stabilized by a kissing-loop interaction (Figure 1. Avsunviroidae) (Bussière et al., 2000, Zhang et al., 2014, Gago et al., 2005) and, as opposed to most viroids, are insoluble in 2 M LiCl (Navarro and Flores 1997). In contrast, eggplant latent viroid (ELVd) and ASBVd fold into quasi-rod-like and rod-like conformations, respectively, and are soluble under the same high salt conditions (Symons 1981, Fadda et al., 2003, López-Carrasco et al., 2016, López-Carrasco and Flores 2017).
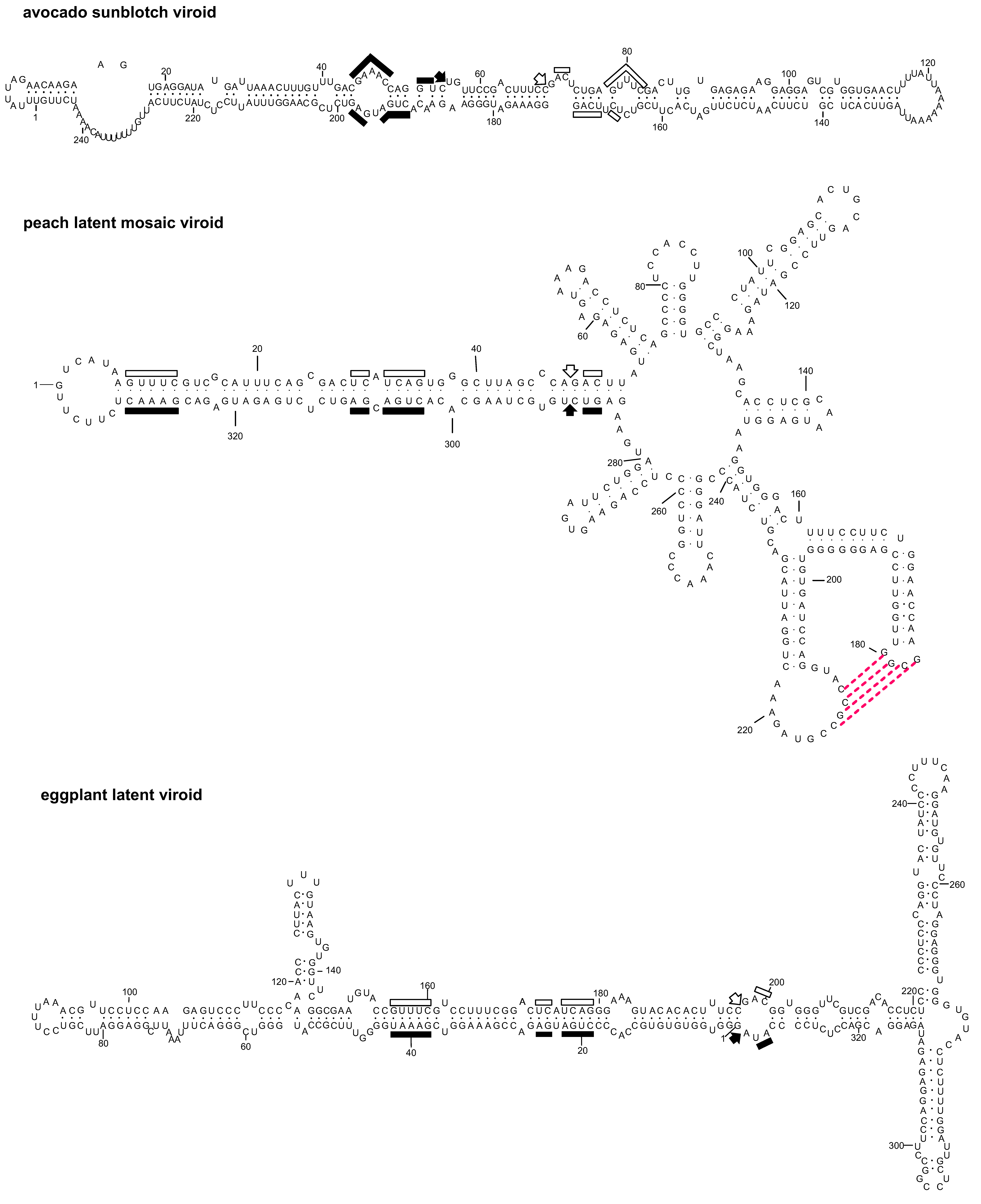 |
| Figure 1. Avsunviroidae. Secondary structure proposed for representative members of the family Avsunviroidae. (Top) Secondary structure of variant X52041, which differs in just one substitution (C123 to U) from the reference variant of the exemplar isolate of avocado sunblotch viroid (J02020, species Avocado sunblotch viroid, genus Avsunviroid) as determined by in vivo SHAPE (López-Carrasco and Flores 2017). (Middle) Secondary structure proposed for the reference variant of the exemplar isolate of peach latent mosaic viroid (M83545, species Peach latent mosaic viroid, genus Pelamoviroid) (Hernández and Flores 1992); a similar structure was confirmed by in vitro SHAPE using another variant of PLMVd (Giguère et al., 2014). (Bottom) Secondary structure of the reference variant of the exemplar isolate of eggplant latent viroid (AJ536613, species Eggplant latent viroid, genus Elaviroid) as determined by in vitro SHAPE (López-Carrasco et al., 2016, Giguère et al., 2014). In the three panels, (+) and (-) sequence motifs conserved in most natural hammerhead structures are indicated by boxes, and self-cleavage sites by arrows. Closed and open boxes indicate the location of the hammerhead structures in the (+) and (-) strands, respectively. In the middle panel, residues involved in a kissing-loop interaction are indicated by red broken lines. |
Nucleic acid
The genome is composed of a circular single-stranded RNA. Strands of (+) and (-) polarity can form hammerheads involved in replication (Figure 2. Avsunviroidae). While the hammerheads of ASBVd are thermodynamically unstable, those of PLMVd, CChMVd. AHVd and ELVd are stable (Fadda et al., 2003, Flores et al., 2000, Serra et al., 2018). Elements of tertiary structure and pathogenic determinants have been identified in several members of the family (de la Peña et al., 1999, Flores et al., 2000, Malfitano et al., 2003).
 |
| Figure 2. Avsunviroidae. Sequence and secondary structure of the hammerhead ribozyme domains of representative members of the family Avsunviroidae. Hammerhead domains of the (+) and (-) polarity (top and bottom panels, respectively) of the reference variant of the exemplar isolate of avocado sunblotch viroid, peach latent mosaic viroid, chrysanthemum chlorotic mottle viroid and eggplant latent viroid. The nucleotides forming the conserved catalytic core are indicated with black background and the self-cleavage sites are marked by arrows; the stems are numbered according to a previous convention (Forster and Symons 1987). (Figure adapted from (Dufour et al., 2009), with permission) While the hammerheads of avocado sunblotch viroid are thermodynamically unstable and have a short helix III, those of peach latent mosaic viroid, chrysanthemum chlorotic mottle viroid and eggplant latent viroid are stable, with the loops capping helices I and II interacting physically via non-canonical interactions. |
Genome organization and replication
Genomic RNAs contain the conserved sequence and structural motifs characteristic of hammerhead ribozymes. These catalytic domains, which can be formed in strands of either polarity (Hutchins et al., 1986), are adopted transiently during transcription but not in the most stable rod-like or branched final conformations, thus avoiding their self-cleavage (Figure 1. Avsunviroidae). Since the circular monomeric RNAs of (+) and (-) polarity accumulate in vivo, a symmetric rolling-circle mechanism (Branch and Robertson 1984, Daròs et al., 1994) has been proposed for their replication that takes place in plastids (Flores et al., 2011) (Figure 1.Viroids). Oligomeric viroid RNAs of both polarities are synthesized by a nuclear-encoded plastid RNA polymerase (NEP) (Navarro et al., 2000, Rodio et al., 2007) that is conscripted to transcribe RNA templates instead of its physiological DNA templates. These oligomers are self-cleaved co-transcriptionally (Carbonell et al., 2006) by the hammerhead ribozymes, which operate as single- or double-structures (Figure 2. Avsunviroidae), thereby generating linear monomeric RNAs that, in turn, are ligated into circular forms by a tRNA ligase (Nohales et al., 2012). This enzyme, like NEP, is encoded in the nucleus and translocated into plastids.
Biology
Host Range
Members of the family Avsunviroidae naturally infect only angiosperms, with a host range restricted to a single plant species or, occasionally, to a few related plant species [reviewed by (Škorić 2017)].
Movement
After entry into a host cell, members of the family Avsunviroidae must be translocated into plastids (mostly chloroplasts), where they replicate and accumulate. Cell-to-cell and systemic spread of these viroids is assumed to take place through plasmodesmata and the phloem, respectively, although conclusive data in this respect are lacking. The pathway involved in viroid trafficking through the double plastid membrane is unknown and essentially no other RNAs in uninfected cells with this ability have been reported. A nuclear step has been proposed for the ELVd transfer into chloroplasts (Gómez and Pallás 2012).
Transmission
Seed transmission has been documented for ASBVd and ELVd (Fadda et al., 2003, Wallace and Drake 1962), but not for PLMVd (Barba et al., 2007, Howell et al., 1998). Instead, transmission to recipient plants through infected pollen has been shown for PLMVd (Barba et al., 2007), while the situation is unclear for ASBVd (Desjardins et al., 1984). Although PLMVd has been experimentally transmitted by Myzus persicae, the relevance of this mode of transmission under natural conditions appears minor (Flores et al., 2017). Horizontal transmission by grafting infected material and sap-contaminated cutting implements occurs frequently in the field, especially in woody hosts. Mechanical transmission in herbaceous hosts is performed by rubbing carborundum-dusted leaves with crude extracts or total nucleic acid preparations from infected tissue, as well as plasmid DNAs containing dimeric head-to-tail cDNA inserts or their in vitro RNA transcripts.
Cross protection and synergism
Superinfection exclusion (cross-protection) between mild and severe strains of members of this family (PLMVd and CChMVd) was reported even before their viroid nature was discovered and used to develop early detection methods (Desvignes 1976, Horst 1975).
Derivation of names
The name of the family Avsunviroidae derives from the type species of the genus Avsunviroid, Avocado sunblotch viroid.
Genus demarcation criteria
The type of hammerhead structure, the G+C content and the solubility in 2 M LiCl, together with clustering in phylogenetic trees derived from the whole sequences, are used to allocate species into genera.
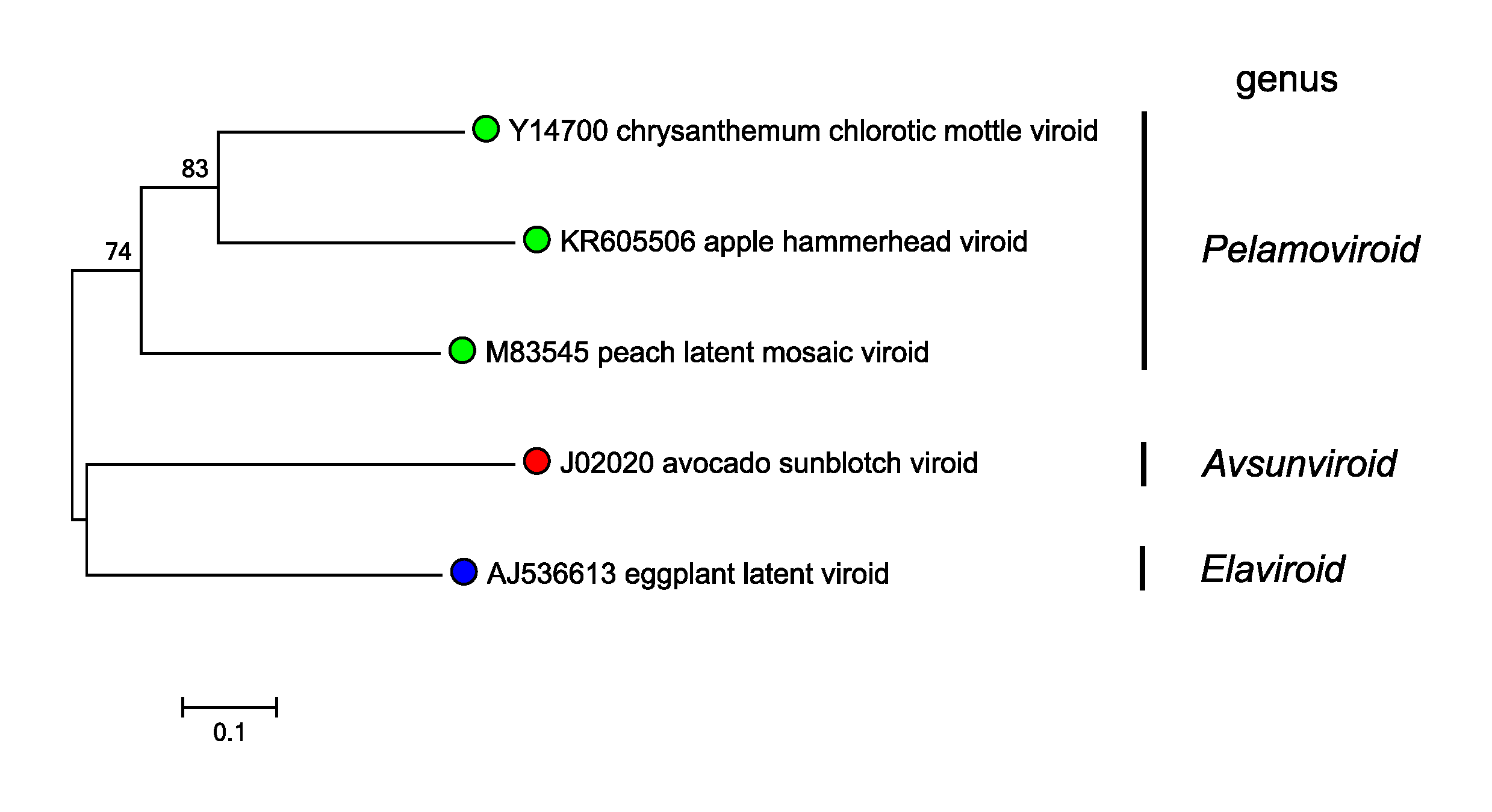 |
| Figure 3. Avsunviroidae. Phylogenetic tree of members of the family Avsunviroidae. Trees were inferred by the neighbor-joining method using MEGA7 (Kumar et al., 2016) with bootstrap values (1000 replicates). Reference sequences for each viroid species were aligned using MUSCLE (position 1 of each sequence corresponds to the nucleotide after the (+) self-cleavage site) with minor manual adjustments. Bootstrap values are reported at the nodes. Evolutionary distances were estimated according to the model of Jukes and Cantor (Jukes and Cantor 1969) eliminating positions with less than 30% coverage. Viroids cluster according to their taxonomic position within the genera in the family. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships within the family
Phylogenetic analysis of whole genome sequences is shown in Figure 3. Avsunviroidae.
Relationships with other taxa
Phylogenetic analysis has related members of the family Avsunviroidae to members of the family Pospiviroidae and to ribozyme-containing viroid-like satellite RNAs (Elena et al., 1991, Elena et al., 2001). Although this relationship has been questioned when tested with alternative phylogenetic approaches (Jenkins et al., 2000), the apparent monophyletic origin of these biologically different infectious circular RNAs supports their possible origin in the RNA world (Diener 1989).
Akin to members of the family Avsunviroidae, certain small circular single-stranded satellite RNAs associated with viruses of the genera Sobemovirus and Polerovirus contain hammerhead ribozymes in strands of both (+) and (-) polarity. However, while viroids replicate autonomously in the host cell, satellite RNAs depend on a co-infecting helper virus (Navarro et al., 2017).
Related, unclassified viroid-like RNAs
A viroid-like RNAs, with hammerhead ribozymes in both polarity strands and adopting branched conformations resembling members of family Avsunviroidae (genus Pelamoviroid), has been reported from grapevine (Wu et al., 2012). Whether it is actually a viroid remains to be confirmed by obtaining data on its independent replication and movement.
| Viroid-like RNA name | Accession number | Abbreviation |
| grapevine hammerhead viroid-like RNA | KR736334 | GHVd-LR |
Virus names and virus abbreviations are not official ICTV designations.

