Family: Virgaviridae
Genus: Pomovirus
Distinguishing features
Pomoviruses have three genomic RNAs, a “triple gene block” set of cell-to-cell movement proteins and are transmitted by root-infecting vectors in the family Plasmodiophorales, once described as fungi but now classified as Cercozoa.
Virion
Morphology
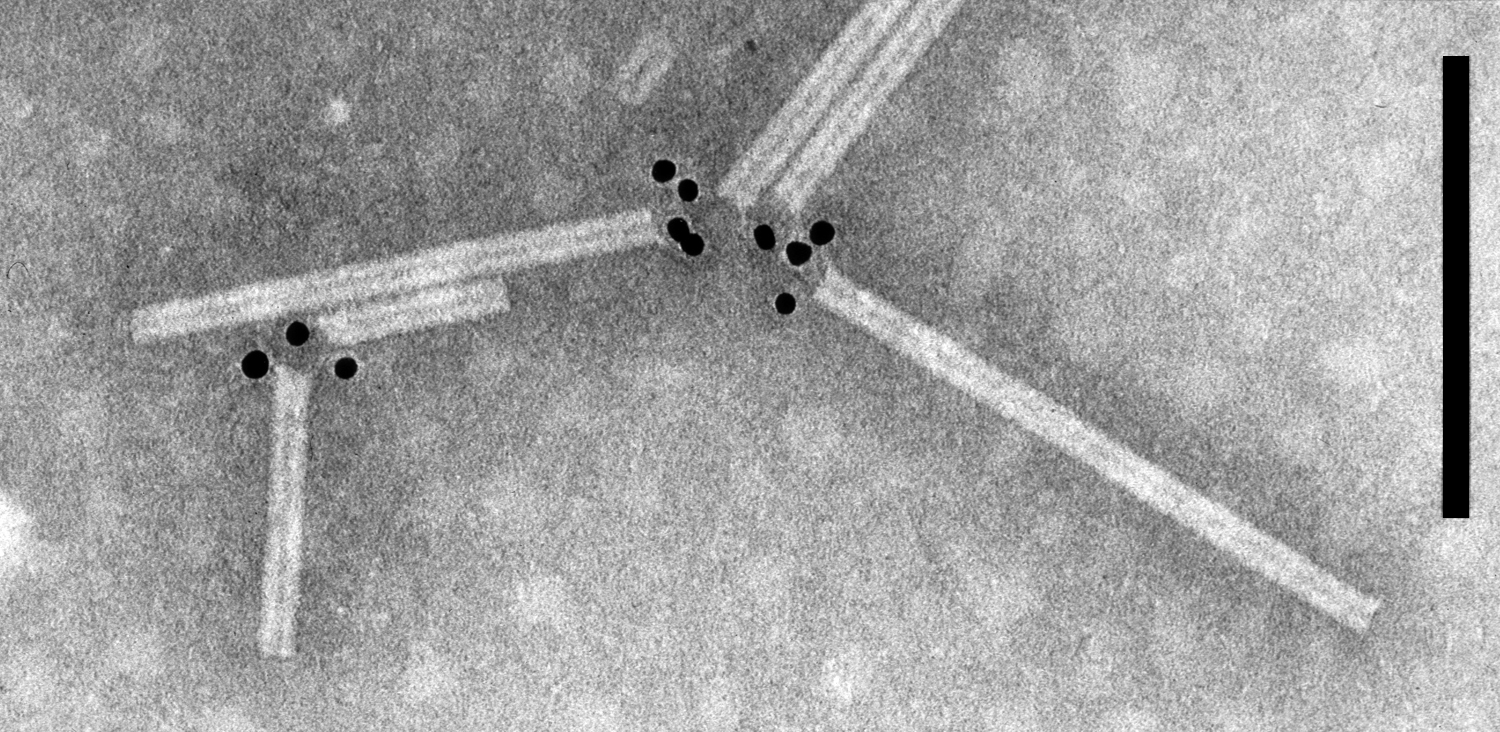
The non-enveloped, rod-shaped particles are helically constructed with a pitch of 2.4 to 2.5 nm and an axial canal (Figure 1. Pomovirus). They have predominant lengths of about 65–80, 150–160 and 290–310 nm and diameters of 18–20 nm. Crude extracts of plants infected with beet soil-borne virus (BSBV), beet virus Q (BVQ) and potato mop-top virus (PMTV) contain characteristic small bundles of side-by-side aggregated particles in addition to singly dispersed particles.
|
Figure 1. Pomovirus. Negative contrast electron micrograph of particles of potato mop-top virus. The gold-labeling shows the binding of monoclonal antibody SCR 68, which detects the CP readthrough protein, to one extremity of the particles. The bar represents 100 nm (Courtesy I.M. Roberts). |
Physicochemical and physical properties
Virions sediment as three components with S20,w of about 125S, 170S and 230S, respectively. In plant sap at room temperature, most virus infectivity is lost within a few hours.
Nucleic acid
Virions contain three molecules of linear positive sense ssRNA of about 6, 3–3.5 and 2.5–3 kb, respectively. The sequence of the three genomic RNAs has been determined for at least one isolate of each species in the genus (Gil et al., 2016, Lu et al., 1998, Koenig et al., 1996, Koenig et al., 1997, Koenig and Loss 1997, Koenig et al., 1998, Sandgren et al., 2001, Savenkov et al., 1999). The RNAs are probably capped at the 5′ end; their 3′ ends can be folded into tRNA-like structures that are preceded by a long hairpin-like structure and an upstream pseudoknot domain. The tRNA-like structures of pomoviruses like those of tymoviruses contain an anticodon for valine and are capable of high-efficiency valylation (Savenkov et al., 1999).
Proteins
The major capsid protein (CP) species is 20 kDa in size. It is not needed for systemic infection. The CP readthrough protein may be detected in some PMTV particles near one extremity by means of immunogold labeling (Cowan et al., 1997). Sequences in the CP readthrough protein are necessary for the transmission of PMTV by Spongospora subterranean (Reavy et al., 1998). Yeast two-hybrid experiments reveal that the CP readthrough protein interacts in vitro with the triple gene block protein movement protein TGB1. In this system, TGB proteins show self interactions and TGB2 and TGB3 interact with each other. TGB2 and TGB3 are membrane-associated and TGB2 binds ssRNA in a sequence nonspecific manner. It has been suggested that they may form a complex with PMTV RNA that is translocated and localized to the plasmodesmata by TGB3 thus facilitating cell-to-cell movement (Tilsner et al., 2010).
Genome organization and replication
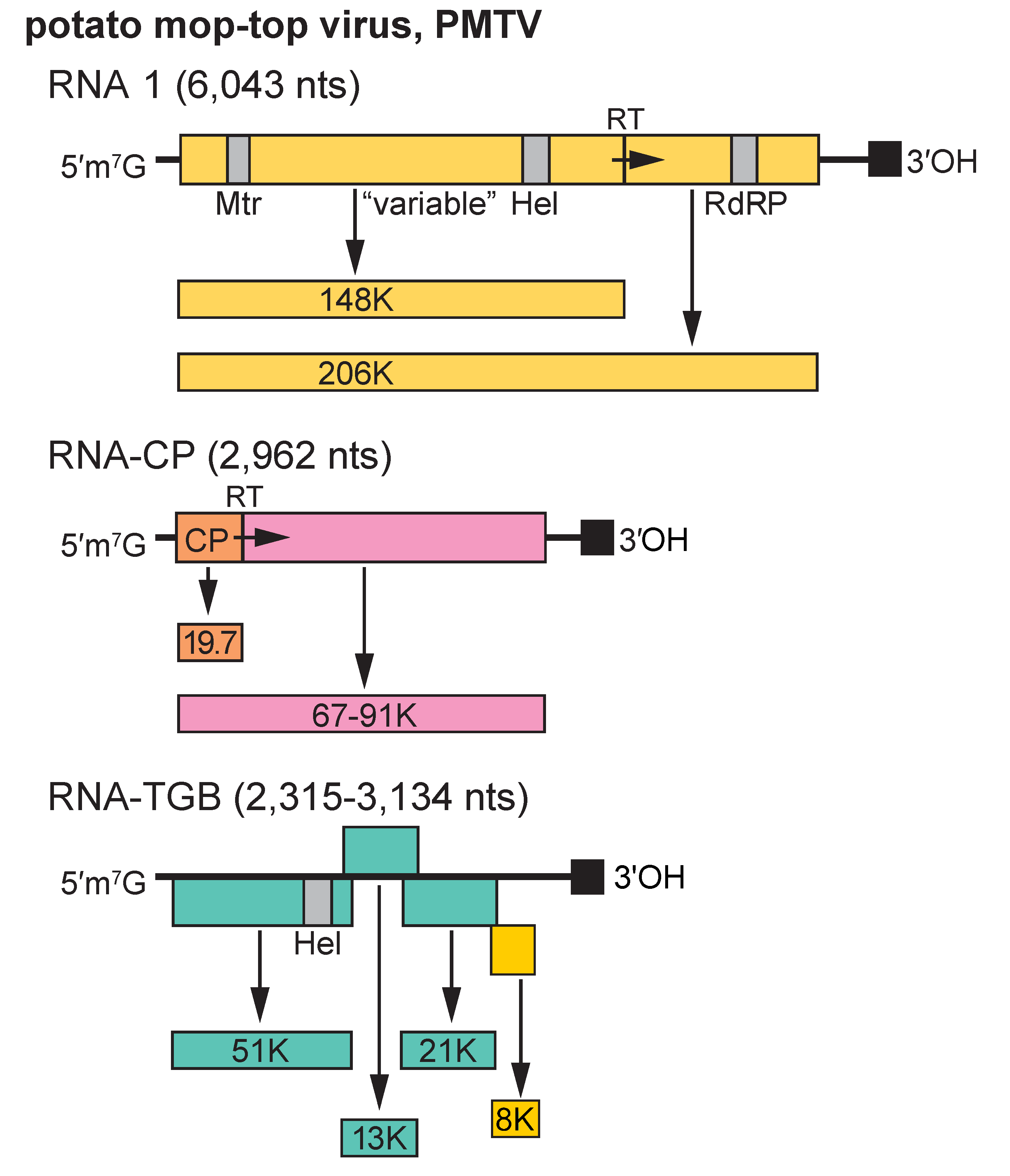
RNA 1 of PMTV has an ORF encoding a 148 kDa protein and a 206 kDa readthrough protein that are presumably involved in replication. The ORF encoding the 148 kDa protein is terminated by an apparently suppressible UGA stop codon (Figure 2. Pomovirus). Proteins of similar sizes are encoded on RNA 1 of BVQ, BBNV and BSBV. The smaller protein contains a Mtr motif in its N-terminal part and a Hel motif in its C-terminal part; the motifs for RdRP are found in the C-terminal part of the readthrough protein (Figure 2. Pomovirus). The two proteins contain other highly conserved domains of unknown function in their N- and C-terminal parts, but their central regions (designated as “variable” in Figure 2. Pomovirus) are specific for each virus. RNA-CP in PMTV (RNA 2 in other viruses), contains the CP ORF, which terminates with a suppressible UAG stop codon and then continues in frame to form a CP readthrough protein that varies in size between different pomoviruses; possibly because it readily undergoes internal deletions. [Note: large deletions have been found in both natural and laboratory isolates of PMTV and PMTV RNA-CP was therefore originally designated as RNA 3]. RNA 3 encodes a triple gene block (TGB) of proteins involved in viral movement; in PMTV there is also a unique 3’-proximal ORF encoding a cysteine-rich protein and in BBNV a predicted 6 kDa glycine-rich protein. TGB1 also contains Hel motifs. The sequences of the C-terminal part of TGB1, of the entire TGB2 and of the N-terminal part of TGB3 are highly conserved among pomoviruses. The replication mechanisms are unknown but both RNA 1 and RNA 3 are essential for BSBV replication and symptom production in Chenopodium quinoa. However, RNA 2 was not essential (Crutzen et al., 2009).
|
Figure 2. Pomovirus. Genome organization typical of potato mop-top virus. Arrows indicate respectively the UGA and UAG stop codons that are thought to be suppressible, and solid squares indicate a 3′-terminal tRNA-like structure. Hel, helicase; Mt, methyltransferase; RdRP, RNA dependent RNA polymerase; RT, readthrough. |
Biology
The natural host range of pomoviruses is very narrow; only dicotyledonous hosts have been described. Pomoviruses are transmitted by soil. Spongospora subterranea and Polymyxa betae have been identified as vectors for PMTV (Arif et al., 1995) and BSBV, respectively. The viruses are also transmissible mechanically.
PMTV-infected cells contain virions aggregated in sheaves in the cytoplasm. Infections with BSBV and BVQ induce voluminous cytoplasmic inclusions which consist of hypertrophied endoplasmic reticulum, convoluted membrane accumulations, numerous small virion bundles and rarely compact virus aggregates.
Antigenicity
Virions are moderately antigenic. Distant serological relationships have been found between the particles of BSBV and BVQ but not between those of the two beet viruses and PMTV. This is probably due to the fact that PMTV CP has an extra ten amino acids on its immunodominant N-terminus that are missing in the CP proteins of the two beet viruses. A conserved sequence EDSALNVAHQL is found in the CPs of PMTV, BSBV and BVQ (Koenig et al., 1998). An epitope within this sequence recognised by the monoclonal antibody SCR 70 is only detectable by Western blotting after disruption of the particles. Other epitopes are either exposed along the entire particle length, e.g. the immunodominant N-terminus, or are accessible only on one extremity (Figure 1. Pomovirus). PMTV and BBNV show distant serological relationships to tobamoviruses.
Derivation of names
Pomo: from potato mop-top virus.
Species demarcation criteria
The criteria demarcating species in the genus are:
- Differences in host range
- Effects in infected tissue: different inclusion body morphology
- Transmission: different vector species
- Serology: virions are distantly related serologically
- Genome: different numbers of genome components (presence or absence of an ORF encoding a cysteine-rich protein)
- Sequence: less than about 80% nt identity over whole genome
- Sequence: less than about 90% identical in CP amino acid sequence
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
Soil-borne virus 2 |
SBV2 |
Virus names and virus abbreviations are not official ICTV designations.
* partial genome