Family: Virgaviridae
Genus: Hordeivirus
Distinguishing features
Hordeiviruses have three genomic RNAs and encode a “triple gene block” set of cell-to-cell movement proteins. They differ from viruses of all other genera because the RdRP is encoded on a separate RNA (rather than by readthrough of a stop codon from an upstream replication protein encoding ORF).
Virion
Morphology
Virions are non-enveloped, elongated and rigid, about 20×110–150 nm in size; they are helically symmetrical with a pitch of 2.5 nm (Figure 1. Hordeivirus). The high-resolution structure of barley stripe mosaic virus (BSMV) obtained by cryo-electron microscopy shows that there are two types of virion that differ in the number of coat protein (CP) subunits per turn, and in the interactions between the CP subunits (Clare et al., 2015).
|
Figure 1. Hordeivirus. Electron micrograph of purified barley stripe mosaic virus (BSMV) particles stained with 2% uranyl acetate. The particles are approximately 20 nm wide and have a length that varies depending on the size of the encapsidated RNA. The field was selected to represent monomers, but often a range of heterodisperse end-to-end aggregates up to 1000 nm in length predominate in purified preparations. The particles in the top left, bottom center, and upper left side of the micrograph are end-to-end aggregates. The bar represents 150 nm. |
Physicochemical and physical properties
BSMV virions occur as heterodisperse sedimenting species with an S20,w of about 182–193S; members of other species in the genus have an S20,w of about 165–200S, depending on the virus. The BSMV isoelectric point is pH 4.5. Thermal inactivation of infectivity occurs at 63–70 °C. Virions are stable and their survival in plant sap ranges from a few days to several weeks.
Nucleic acid
Virions normally contain three positive sense ssRNAs. The RNAs are designated α (RNA 1), β (RNA 2) and γ (RNA 3), and their respective sizes are 3.8, 3.2 and 2.8 kb (BSMV-ND18 strain), 3.8, 3.0 and 2.7 kb (Lychnis ringspot virus, LRSV), and 3.8, 3.6 and 3.1 kb (Poa semilatent virus, PSLV). The sizes of the α and β RNAs are similar between different strains of BSMV, whilst RNAγ varies in size because of internal duplications of unknown significance.
Each RNA has m7GpppGUA at its 5′ end, and a highly conserved 238 nt (BSMV), 184 nt (LRSV), or 268 nt (PSLV) tRNA-like structure at the 3′ end. In the case of BSMV, this structure can be charged with tyrosine. In the BSMV and LRSV genomes, a poly(A) sequence that is variable in length (~20 nt in BSMV, ~30-50 nt in LRSV) separates the coding region from the tRNA-like structure; however, this sequence is not present in the PSLV genome. A close sequence similarity between the first 70 nt of RNAα and RNAγ of the CV17 strain of BSMV suggests that a natural recombination event has occurred. A similar recombination event appears to have occurred between the 5′-untranslated leaders of RNAα and RNAβ of LRSV. These findings, plus sequence duplications in RNAγ, provide persuasive evidence that RNA recombination has had a substantial role in the evolution of hordeiviruses (Edwards et al., 1992).
Proteins
The virion capsid is constructed from subunits of a single protein. The CP of all species is 22 kDa in size, yet the proteins differ in electrophoretic mobility.
Genome organization and replication
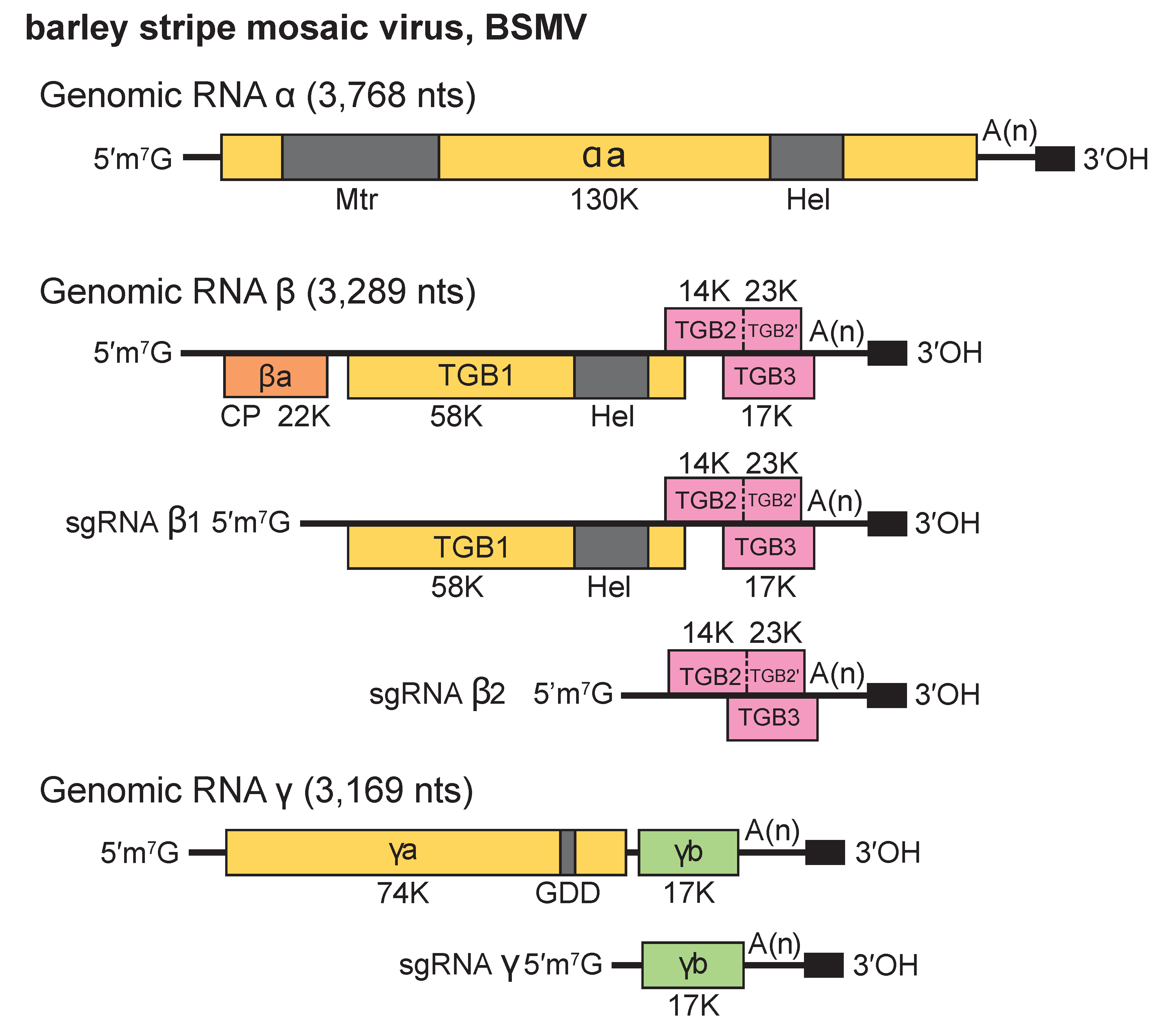
All three BSMV genomic RNAs are required for systemic infection of plants, but RNAs α and γ alone can infect protoplasts. The 5′- and 3′- non-coding regions (NCR) of each BSMV RNA are required for replication (Jackson et al., 2009). The hordeivirus genome encodes seven proteins as illustrated for BSMV in Figure 2. Hordeivirus.
|
Figure 2. Hordeivirus. Genome organization of barley stripe mosaic virus (BSMV). The colored rectangles and smaller solid black rectangles represent the ORFs, and the 3′-terminal tRNA-like structure, respectively. The 3′-proximal ORFs on each RNA terminate with an UAA that initiates the short poly (A) tract directly preceding the 238 nt tRNA-like terminus. See text for explanation. |
RNAα is monocistronic and encodes the αa protein (130 kDa in BSMV, 129 kDa in LRSV and 127 kDa in PSLV) that functions as the helicase subunit of the viral replicase. The αa protein has two conserved sequence domains, an amino-terminal Mtr and a carboxy-terminal NTPase/Hel.
The 5′-terminal ORFs of RNAβ (βa) of BSMV, LRSV and PSLV encode a 22 kDa CP. The BSMV CP, which is dispensable for systemic movement of the virus, is more closely related to the PSLV CP (53.2% aa identity) than to the LRSV CP (33.7% aa identity). An intergenic region separates a “triple gene block” (TGB) that encodes three nonstructural proteins, TGB1, TGB2 and TGB3. In BSMV, The TGB1 protein is expressed from a 2,450 nt subgenomic RNA (sgRNAß1) , and the TGB2 and TGB3 proteins are expressed from a second bicistronic 960 nt subgenomic RNA (sgRNA ß2) with TGB3 being expressed via a leaky scanning mechanism. In BSMV, a minor 23 kDa translational readthrough extension of the TGB2 protein, designated TGB2′, is present in plants. However, genetic experiments have not identified a function for TGB2′, so it appears to be dispensable for infection in all local lesion and systemic hosts tested. The BSMV sgRNAβ1 and sgRNAβ2 promoters reside between positions −29 to −2 and −32 to −17 relative to the transcription initiation sites, respectively, and the nt sequences preceding the transcription initiation sites of these sgRNAs are conserved in LRSV and PSLV. The TGB1 protein (58 kDa in BSMV, 50 kDa in LRSV, and 63 kDa in PSLV) contains a conserved NTPase/Hel domain. The BSMV TGB1 protein binds RNA, NTPs and exhibits ATPase and helicase activity in vitro (Donald et al., 1997). It also elicits resistance to BSMV strains that are unable to infect Brachypodium distachyon inbred lines containing the Bsr1 resistance gene. Protein kinase CK2 phosphorylation of the TGB1 protein plays a critical role in promoting viral movement in monocots and dicots by affecting the interactions between the TGB1 and TGB3 proteins (Lee et al., 2012, Hu et al., 2015). The TGB2 (17 kDa in BSMV, 14 kDa in LRSV and 15 kDa in PSLV) and TGB3 (14 kDa in BSMV, 18 kDa in LRSV, and PSLV) proteins are hydrophobic and membrane-associated. Each of the BSMV TGB proteins is required for virus cell-to-cell movement in plants.
RNAγ is bicistronic and encodes the γa polymerase subunit of the viral replicase (74 kDa in the BSMV-ND18 strain, 72 kDa in LRSV, and 81 kDa in PSLV), and the cysteine-rich γb protein (17 kDa in BSMV, 16 kDa in LRSV, and 20 kDa in PSLV). The γa protein is variable in size because of a repeated sequence of 351-363 nt present in some strains of BSMV (Kozlov Iu et al., 1989). The BSMV γb protein is expressed from a 737 nt subgenomic RNA (sgRNAγ) and is a pathogenicity determinant involved in regulating expression of ORFs encoded by RNAβ. The sgRNAγ promoter is between nt −21 to +2 relative to its transcription start site, and this sequence has similarity to sequences upstream of the γb proteins in PSLV and LRSV. The BSMV γb protein has both RNA binding and zinc binding ability, participates in homologous interactions, and may act as a suppressor of post-transcriptional gene silencing.
Translation of a functional αa protein is required for replication of RNAα in cis, whilst replication of RNAβ is dependent on the presence of the CP and TGB1 intergenic region, and replication of RNAγ depends upon approximately 600 nt of the 5′-terminal region. The TGB proteins encoded on RNAβ are required for cell-to-cell and systemic movement in plants, but the CP and TGB2′ are dispensable. The γb protein is also dispensable in some genetic backgrounds. A mutation in the 5′-NCR sequence of the γa ORF interfered with systemic infection of Nicotiana benthamiana, suggesting that modulation of γa expression can affect movement. Full-length dsRNAs corresponding to all viral genomic ssRNAs can be isolated from infected plants. Virus particles accumulate predominantly in the cytoplasm and also in nuclei. Infected barley plants develop pronounced enlargements of the plasmodesmata that contain the TGB1 protein (Gorshkova et al., 2003), and prominent peripheral vesicles appear in proplastids and chloroplasts. These vesicles are the sites of replication (Torrance et al., 2006, Zhang et al., 2017).
Biology
The native hosts of three viruses (ALBV, BSMV, PSLV) are grasses (family Gramineae); strains of LRSV occur naturally in dicotyledonous plants of the families Caryophyllaceae and Labiatae. Various strains of these viruses elicit local lesions in Chenopodium species and are able to establish systemic infections in a common host, Nicotiana benthamiana. BSMV and LRSV are efficiently seed-transmitted and are transmitted less efficiently by pollen. Field spread from primary infection foci occurs efficiently by direct leaf contact. There are no known vectors for any members of the genus. Anthoxanthum latent blanching virus (ALBV) has been reported only from Great Britain; BSMV occurs world-wide wherever barley is grown and has recently been isolated from 750-year-old barley grains found near the Nile River (Smith et al., 2014); LRSV (mentha strain) has been isolated in Hungary (Beczner et al., 1992), and the type strain which is highly seed-transmissible in the family Caryophyllaceae, was initially discovered in California from seed of Lychnis divaricata introduced from Europe. PSLV has been recovered from Poa palustris isolated from two locations in Western Canada (Slykhuis 1972).
Antigenicity
Hordeivirus particles are strong immunogens. Member species are distantly related serologically with BSMV being more closely related to PSLV than to LRSV, which is in agreement with sequence analyses.
Derivation of names
Hordei: from hordeus, Latin name of the primary host of the type species virus of the genus Hordeivirus.
Species demarcation criteria
Species differ in host range and are phylogenetically distinct. Precise molecular discrimination criteria have not been established because, except for isolates of the type species, few sequences have been determined.