Family: Virgaviridae
Genus: Pecluvirus
Distinguishing features
Pecluviruses have a bipartite genome, a “triple gene block” set of cell-to-cell movement proteins and are transmitted by root-infecting vectors in the family Plasmodiphorales, once described as fungi but now classified as Cercozoa.
Virion
Morphology
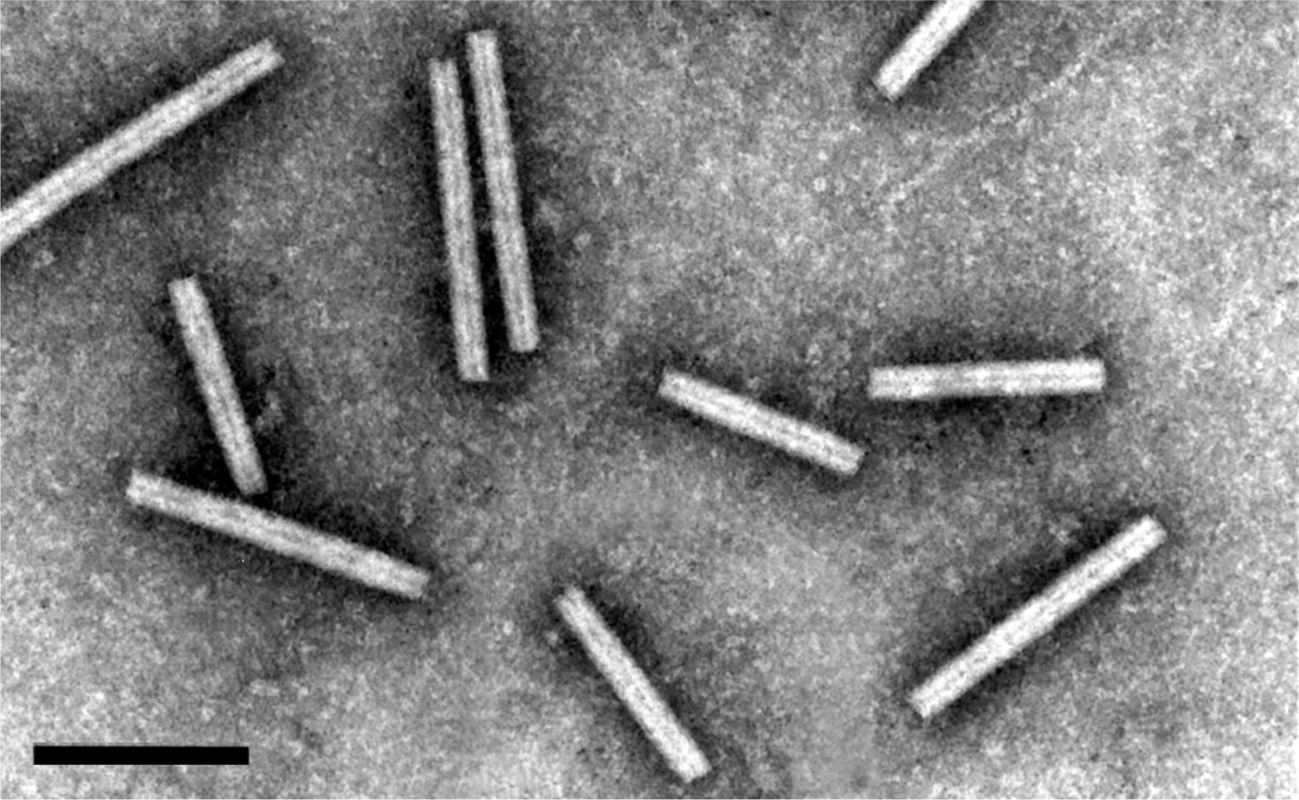
Virions are rod-shaped, about 21 nm in diameter and of two predominant lengths, 190 and 245 nm (Figure 1.Pecluvirus). The length distribution of the short particles is broad and in some preparations an additional class of 160 nm particles is recognizable. Virions have helical symmetry with a pitch of 2.6 nm.
|
Figure 1. Pecluvirus. Negative contrast electron micrograph of virions of Indian peanut clump virus (L serotype) negatively stained with 2% phosphotungstic acid, pH 6. The bar represents 150 nm. |
Physicochemical and physical properties
Virions sediment as two major components with S20,w of 183S and 224S. Buoyant density in CsCl is 1.32 g cm−3. Virion isoelectric point is pH 6.45. Thermal inactivation of infectivity occurs at 64 °C. Virions are stable in frozen leaves.
Nucleic acid
The genome consists of two molecules of linear positive sense ssRNA: RNA 1 of about 5,900 nt and RNA 2 of about 4,500 nt. RNAs are thought to have a 5′-cap structure but this has not been confirmed. The 3′ ends of the RNAs can fold into a tRNA-like structures and are not polyadenylated.
Proteins
The virion CP proteins are 23 kDa.
Genome organization and replication
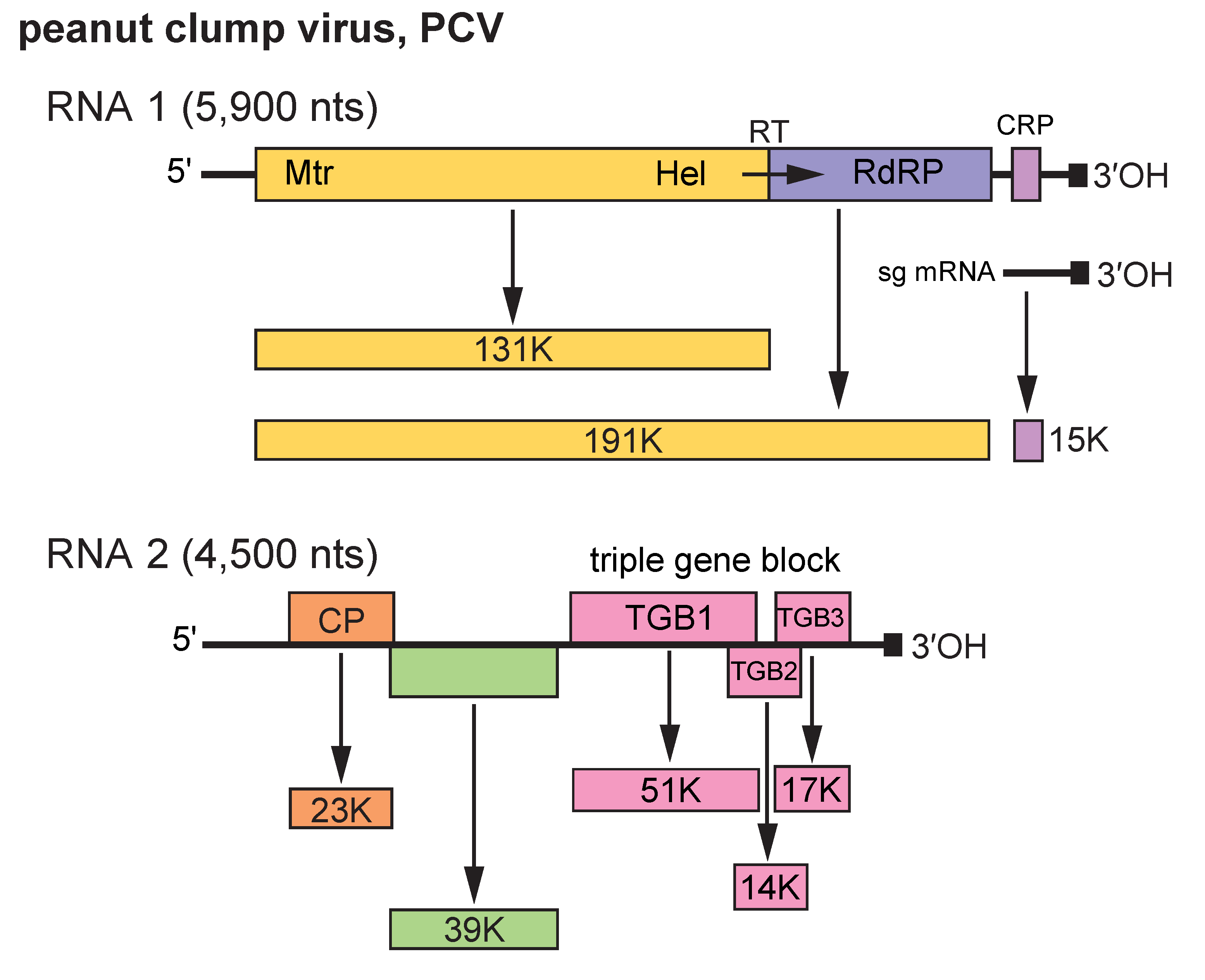
RNA 1 contains two ORFs (Figure 2.Pecluvirus). The 5′-proximal ORF encodes a 131 kDa protein and suppression of a termination codon can result in the synthesis of a readthrough protein of 191 kDa. The 3′ proximal ORF encodes a 15 kDa protein. The proteins of 131 and 191 kDa contain NTP-binding, Hel and RNA polymerase motifs and are involved in the putative replication complex (Miller et al., 1996, Herzog et al., 1994). The 15 kDa protein is translated from a subgenomic RNA. It is a suppressor of post-transcriptional gene silencing (Dunoyer et al., 2002a) and is targeted to peroxisomes or related punctate bodies during infection. RNA 2 contains five ORFs (Manohar et al., 1993): the 5′ proximal ORF encodes the CP, the adjacent ORF which, in PCV RNA 1, overlaps the first ORF by 2 nts encodes a 39 kDa protein. This protein is expressed by leaky scanning (Herzog et al., 1995) and is thought to be involved in the transmission of PCV by its fungus vector. Further downstream, and separated by a 135 nt intergenic region, is a triple gene block sequence that codes for proteins of 51, 14 and 17 kDa that are thought to be involved in the movement of virus from cell to cell. These proteins are probably expressed via one or more sgRNAs but these have not been clearly identified. The 3′-NCRs for PCV are 298 nt for RNA 1 and 275 nt for RNA 2; the 3' terminal 96 nt are identical in both RNAs. The NCRs differ in size among isolates from the different serotypes of IPCV.
Both genomic RNAs are required for systemic invasion of plants but RNA 1 is able to replicate in absence of RNA 2 in protoplasts (Dunoyer et al., 2002b). The virus is found in the cells of roots, stems and leaves of systemically infected plants.
|
Figure 2. Pecluvirus. Genomic organization of peanut clump virus (PCV). The tRNA structure motifs at the 3′-ends of the RNAs are represented by a dark square. ORFs are indicated by rectangles and a suppressible termination codon by an arrow (RT, readthrough). CRP, cysteine-rich protein. |
Biology
The first reported natural host of pecluvirus was Arachis hypogea (groundnut, Leguminosae). Disease symptoms are stunting – mottle – mosaic – chlorotic ringspot. PCV infects Sorghum arundinaceum, usually symptomlessly. IPCV infects a number of cereal crops and graminaceous weeds, some symptomlessly and others to induce stunting. The experimental host range is wide and includes species of Aizoaceae, Amaranthaceae, Chenopodiaceae, Cucurbitaceae, Graminae, Leguminosae, Scrophulariaceae and Solanaceae. Nicotiana benthamiana and Phaseolus vulgaris are experimental propagation hosts. Chenopodium amaranticolor and Chenopodium quinoa are local lesions hosts (Thouvenel and Fauquet 1981).
The virus is transmitted naturally by Polymyxa graminis or by seed (in groundnuts) (Dieryck et al., 2011, Dieryck et al., 2009). It is mechanically transmissible.
PCV exists in West Africa (Bénin, Burkina Faso, Congo, Côte d’Ivoire, Mali, Niger, Senegal and Pakistan). IPCV is widely distributed in India and Pakistan. A soil type favourable to the vector is a prerequisite for the virus to cause disease.
Antigenicity
The viruses are highly immunogenic. There is a serological variability among isolates of PCV. IPCV isolates fall into one of three very distinct serotypes: IPCV-H, IPCV-L, IPCV-T (Huguenot et al., 1989). All are serologically distinct from PCV.
Derivation of names
Peclu: from peanut clump virus.
Species demarcation criteria
There are two species in the genus. Isolates of both species are serologically distinct (heterologous reactions are weak or undetectable) and differ by geographical distribution (PCV only in Africa; IPCV in the Indian subcontinent). There are also serologically distinct strains within each species and better demarcation criteria may need to be re-examined when more isolates have been studied or other members of the genus are discovered.