Family: Virgaviridae
Genus: Furovirus
Distinguishing features
Furoviruses have a bipartite genome, a “30K”-like cell-to-cell movement protein and are transmitted by root-infecting vectors in the family Plasmodiphorales, once described as fungi but now classified as Cercozoa.
Virion
Morphology
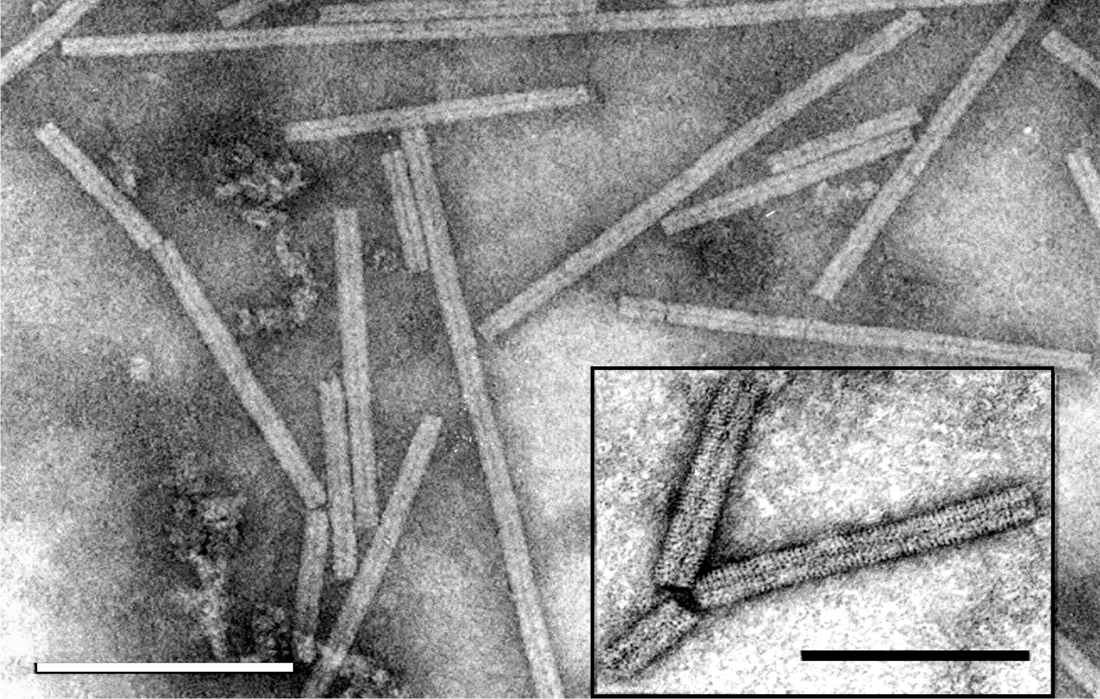
Virions are non-enveloped hollow rods, which have helical symmetry. Virions are about 20 nm in diameter, with predominant lengths of 140–160 nm and 260–300 nm. The length distribution of the soil-borne wheat mosaic virus (SBWMV) short particles is broad, 80–160 nm, due to the presence of deletion mutants in some cultures (Figure 1. Furovirus).
|
Figure 1. Furovirus. Negative contrast electron micrograph of stained (ammonium molybdate pH 7.0) particles of soil-borne wheat mosaic virus (SBWMV). The bar represents 200 nm. Inset: Negative contrast electron micrograph of particles SBWMV stained with 1% uranyl acetate. The bar represents 100 nm. |
Physicochemical and physical properties
Virions sediment as two (or three) components; for SBWMV the S20,w values are 220–230S (long particles) and 170–225S (short particles), and 126–177S (deletion mutants). SBWMV loses infectivity in extracts of wheat kept at 60–65 °C for 10 min.
Nucleic acid
Complete or almost complete sequences are available for isolates representing all species in the genus (Shirako and Wilson 1993, Diao et al., 1999a, Shirako et al., 2000, Diao et al., 1999b). The genome is bipartite, linear, positive sense ssRNA. RNA 1 is about 6–7 kb and RNA 2 about 3.5–3.6 kb. The RNA molecules of SBWMV have a 5′ cap (m7GpppG) and in all of the species where the complete sequences have been determined there is a 3′-terminal tRNA-like structure with a putative anti-codon for valine. The 3′ terminus of SBWMV RNA was shown experimentally to accept valine (Goodwin and Dreher 1998).
Proteins
The capsid comprises (or mostly comprises) multiple copies of a polypeptide of about 19–20.5 kDa. The CPs of SBWMV, Chinese wheat mosaic virus (CWMV), soil-borne cereal mosaic virus (SBCMV) and oat golden stripe virus (OGSV) comprise 176 aa with 76–82% aa identity; they share only 46% identity with that of sorghum chlorotic spot virus (SrCSV). The CP ORF terminates in a leaky (UGA) stop codon that can be suppressed to produce a read-through minor capsid protein (ca. 85 kDa), which is thought to be involved in natural transmission by the plasmodiophorid vector. In SBWMV and CWMV, it has been experimentally shown that a further minor coat protein of 25 kDa is initiated from a CUG codon upstream of the canonical CP AUG but in SBWMV neither minor capsid protein was essential for particle formation or systemic infection (Shirako 1998, Yamamiya and Shirako 2000, Yang et al., 2016).
Genome organization and replication
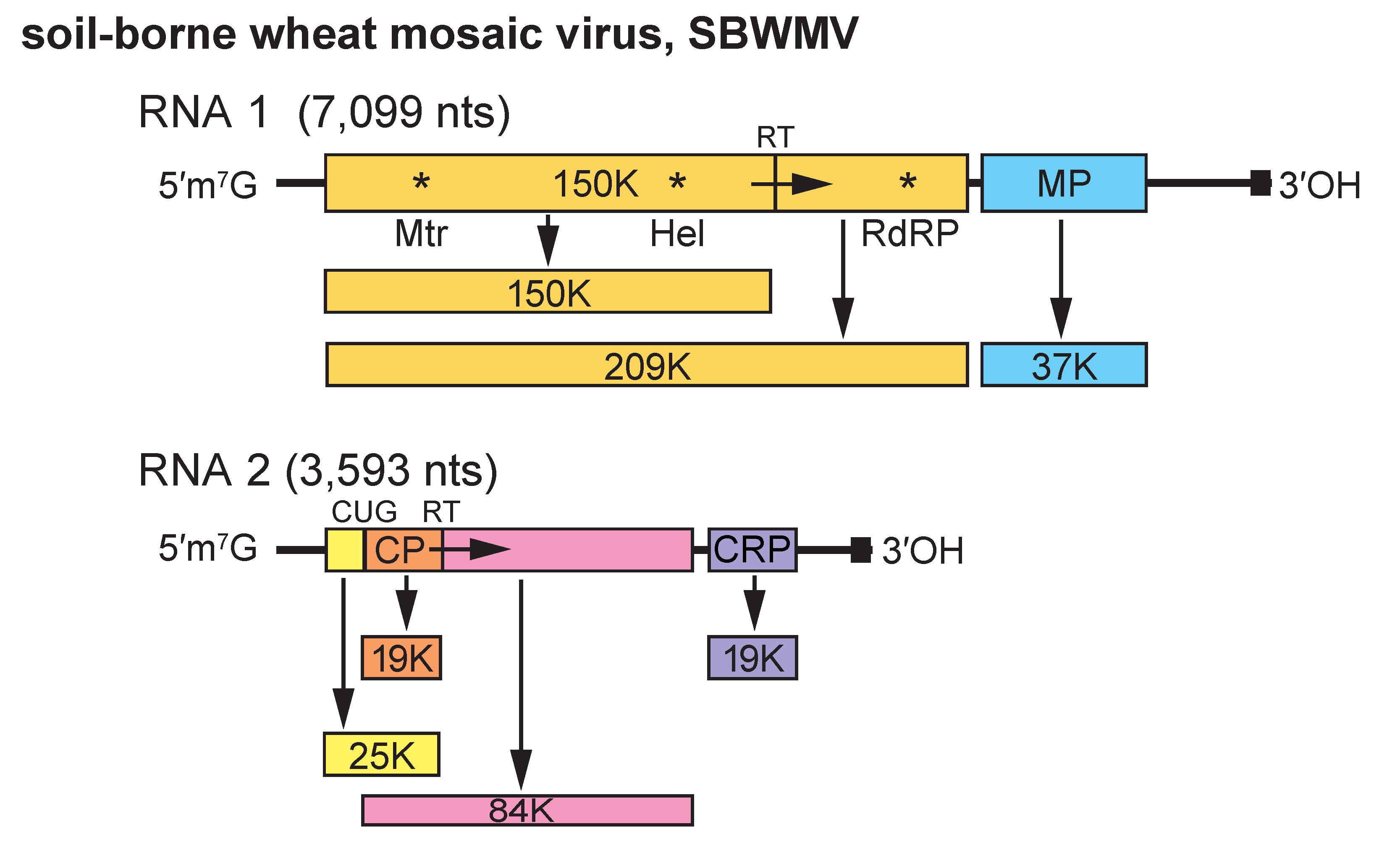
Genome organization and structure are conserved between members of the species but there are substantial differences in the genomic sequences. SBWMV RNA 1 encodes a 150 kDa protein, a 209 kDa readthrough product and a 37 kDa protein (Figure 2. Furovirus). The 150 kDa protein contains Mtr and NTP-binding Hel motifs. The readthrough protein, in addition, contains RNA polymerase motifs, indicating that these proteins are involved in replication. The 37 kDa protein belongs to the “30K”-like cell-to-cell movement protein superfamily and is thought to be involved in virus movement as it shares some sequence similarity to the MPs of dianthoviruses (An et al., 2003). RNA 2 encodes the CP (19 kDa), the sequence of which terminates in a UGA codon that can be suppressed to give a readthrough product of 84 kDa. Also, a 25 kDa polypeptide is initiated from a CUG codon upstream of the CP AUG. A 3′ proximal ORF of RNA 2 encodes a 19 kDa protein that contains seven conserved cysteine residues. This protein is a suppressor of gene silencing (Te et al., 2005, Sun et al., 2013). Products corresponding to the 37 kDa protein and the cysteine-rich 19 kDa protein were not found in in vitro transcription/translation experiments, and these proteins are thought to be expressed from subgenomic mRNAs. Spontaneous deletions in RNA2 occur on successive passage by manual inoculation, and in field isolates in older infected plants, resulting in a shorter CP readthrough product (Chen et al., 1994, Chen et al., 1995).
|
Figure 2. Furovirus. Genome organization of soil-borne wheat mosaic virus (SBWMV). The tRNA structure motifs at the 3′-ends of the RNAs are represented by a dark square, the methyl transferase (Met), Helicase (Hel) and RNA-dependent RNA polymerase (RdRP) motifs by asterisks and the readthrough of the polymerase and coat protein ORFs by RT and an arrow. The major CP (19K) is shown in orange. A minor 25K CP is initiated from an upstream CUG codon. CRP, cysteine-rich protein. |
Biology
Furoviruses are found in temperate regions worldwide including the United States of America, Europe, China and Japan. The natural host ranges of furoviruses are narrow and confined to species within the Graminae. SBWMV induces green or yellow mosaic and stunting in winter wheat (Triticum aestivum) causing up to 80% yield loss in severely infected crops. It also may infect barley and rye. SBCMV infects mainly wheat and triticale in Western and Southern Europe and mainly rye in Central and North-Eastern Europe. Both viruses are (not readily) mechanically transmissible to Chenopodium quinoa. OGSV infects oats (Avena sativa) but failed to infect wheat when plants were grown in viruliferous soil. Mechanically OGSV can be transmitted to some Nicotiana and Chenopodium species. SrCSV infects Sorghum bicolor and can be mechanically transmitted to a range of species including Chenopodium quinoa, C. amaranticolor, Nicotiana clevelandii, Arachis hypogaea, Zea mays and T. aestivum.
The furoviruses are soil-borne; Polymyxa graminis has been identified as a vector for SBWMV (Estes and Brakke 1966) and is probably the vector of the other viruses in the genus. Virions are thought to be carried within the motile zoospores. Soil containing the resting spores remains infectious for many years.
Virions are found scattered, or in aggregates and inclusion bodies in the cytoplasm and vacuole. Inclusion bodies can be crystalline inclusions or comprise loose clusters of virus particles in association with masses of microtubules. Amorphous inclusion bodies can be seen in tissue sections by light microscopy (Peterson 1970, Hibino et al., 1974a, Hibino et al., 1974b).
Antigenicity
Virions are immunogenic and members of the different species can be distinguished serologically.
Derivation of names
Furo: from fungus-borne, rod-shaped virus.
Species demarcation criteria
Isolates of different species have less than 75% nt identity for RNA 1 and less than 80% nt identity for RNA 2. SBWMV, SBCMV, CWMV and OGSV can be discriminated also by reactivity with selected monoclonal and polyclonal antibodies. OGSV and SrCSV differ in host range to SBWMV, SBCMV and CWMV. The latter three viruses have similar biological properties and experimental re-assortants of SBWMV (Nebraska), SBWMV (Japan) and SBCMV were infectious (Miyanishi et al., 2002). With the other viruses this possibility has not yet been checked.