Family: Virgaviridae
Michael J. Adams, Scott Adkins, Claude Bragard, David Gilmer, Dawei Li, Stuart A. MacFarlane, Sek-Man Wong, Ulrich Melcher, Claudio Ratti and Ki Hyun Ryu
Corresponding author: Michael J Adams (mike.adams.ictv@gmail.com)
Edited by: Hélène Sanfaçon and Stuart G. Siddell
Posted: August 2017
PDF: ICTV_Virgaviridae.pdf
Summary
The Virgaviridae is a family of plant viruses with rod-shaped virions, a single-stranded RNA genome with a 3′-terminal tRNA-like structure and a replication protein similar to those of the alpha-like supergroup (Table 1. Virgaviridae). Differences in the numbers of genome components, genome organisation and the modes of transmission provide the basis for genus demarcation.
Table 1. Virgaviridae. Characteristics of members of the family Virgaviridae.
| Characteristic | Description |
| Typical member | tobacco mosaic virus variant 1 (V01408), species Tobamovirus tabaci |
| Virion | Non-enveloped, rod shaped particles about 20 nm in diameter and up to 300 nm long. Except in members of the genus Tobamovirus, the particle length distribution is bi- or tri-modal |
| Genome | Approximately 6.3 to 13 kb of positive-sense RNA; non-segmented in members of the genus Tobamovirus but multipartite in other genera with segments separately encapsidated in 2 or 3 components |
| Replication | Cytoplasmic, probably associated with the endoplasmic reticulum |
| Translation | From genomic or subgenomic RNAs |
| Host range | Plants (all genera); furoviruses, peculviruses and pomoviruses are transmitted by plasmodiophorids, tobraviruses are transmitted by nematodes and goraviruses and hordeiviruses are transmitted by pollen and/or seed. Tobamoviruses have no known vectors but are readily transmitted mechanically |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Martellivirales; 7 genera containing 59 species |
Viruses classified into the seven genera show distinct host ranges, genome organisation and modes of transmission:
Goravirus. Members of the type species (Gentian ovary ringspot virus, GORV) of this genus are transmitted through pollen, so resembling hordeiviruses. Genomes are bipartite and encode "triple gene block" (TGB) movement proteins, thus resembling pecluviruses but there are differences between members of the two genera in the position of some other open reading frames (ORFs).
Furovirus. This genus contains viruses of graminaceous plants that are transmitted to plant roots by the plasmodiophorid Polymyxa graminis. Genomes are bipartite and encode a ‘30K’-type movement protein. Soil-borne wheat mosaic virus and similar viruses cause serious diseases of winter cereals in North America, Europe and Asia.
Hordeivirus. This genus contains viruses of graminaceous plants that are transmitted through pollen and seed. The genomes are tripartite and (uniquely within the family), the RNA-dependent RNA polymerase is encoded on a separate RNA segment and not as a readthrough product translated together with the other viral-encoded replication proteins. Barley stripe mosaic virus has a worldwide distribution.
Pecluvirus. This genus contains soil-borne viruses of leguminous and graminaceous plants from India and Africa that are transmitted to plant roots by the plasmodiophorid Polymyxa graminis. Genomes are bipartite; they encode TGB movement proteins.
Pomovirus. This genus contains viruses of solanaceous and chenopodiaceous plants that are transmitted to plant roots by the plasmodiophorids Spongospora subterranea and Polymyxa betae. Genomes are tripartite; they encode TGB movement proteins and the coat protein has a carobxyl (readthrough) extension associated with transmission.
Tobamovirus. This genus contains viruses with monopartite genomes that encode a ‘30K’-type movement protein. Tobacco mosaic virus was the first virus discovered (in 1886); it is present in high concentrations in infected plants, is extremely stable, and has been extensively studied (Harrison and Wilson 1999, Scholthof 2004).
Tobravirus. This genus contains viruses of solanaceous plants that are transmitted to plant roots by nematodes. Genomes are bipartite and they encode a ‘30K’-type movement protein.
Virion
Morphology
The non-enveloped, rod-shaped particles are helically constructed with a pitch of 2.3 to 2.5 nm and an axial canal. They are about 20 nm in diameter, with predominant lengths that depend upon the genus.
Physicochemical and physical properties
The S20,w values range from 194 to 306 for large particles that include RNA encoding the replication protein and 125 to 245 for smaller particles. Particles are stable at higher temperatures (60–90 °C) with the exception of pomoviruses, which lose infectivity at room temperature within a few hours.
Nucleic acid
The genome consists of positive sense ssRNA with 5′-cap (m7GpppG) and a 3′-terminal tRNA-like structure. The number of genome components depends upon the genus.
Proteins
The capsid comprises multiple copies of a single polypeptide of about 17–24 kDa, depending upon the genus. Some viruses encode an additional capsid protein (CP) produced by suppression of the CP ORF stop codon to produce a larger readthrough (RT) protein of variable mass called the minor CP or CP-RT.
Genome organization and replication
The largest ORF encodes an alpha-like replication protein with conserved methyltransferase (Mtr) and helicase (Hel) domains. This protein is translated directly from genomic RNA. In viruses of all genera except Hordeivirus, the RNA-dependent RNA polymerase (RdRP) is expressed as the C-terminal part of this protein by readthrough of a leaky stop codon. Other ORFs are expressed either directly from the smaller genomic RNAs or from subgenomic mRNAs, some of which may be bicistronic. In some genera, the viruses have a single cell-to-cell movement protein (MP) of the “30K” superfamily (Melcher 2000), while in other genera there is a triple gene block (TGB). There are differences in the number of genomic RNAs (1, 2 or 3 depending on the genus). Replication is cytoplasmic.
Biology
Biologically, the viruses are fairly diverse. They have been reported from a wide range of herbaceous and mono- and dicotyledonous plant species but the host range of individual members is usually limited. All members can be transmitted experimentally by mechanical inoculation, and for those in the genus Tobamovirus, this is the only known means of transmission. In some genera, transmission is by soil-borne vectors, while members of the genus Hordeivirus and Goravirus are transmitted through pollen and seed.
Antigenicity
Virions are moderately to strongly antigenic.
Derivation of names
Virga: from the Latin for ‘rod’, referring to the morphology of virus particles.
Genus demarcation criteria
Genera are distinguished by the number of genomic RNAs, various features of genome organization, the type of cell-to-cell movement protein and the natural mode of transmission. These are summarized in Table 2. Virgaviridae. Genus distinctions are also supported by phylogenetic analyses of homologous proteins.
Table 2. Virgaviridae. Distinguishing properties of genera in the family Virgaviridae.
| Genus | RNAs | RdRPa | MPb | CPc | 3′ structured | Transmission |
|---|---|---|---|---|---|---|
| Goravirus | 2 | RT | TGB | 22K | t-RNA? | pollen |
| Furovirus | 2 | RT | “30K” | 19K+RT | t-RNAVal | plasmodiophorid |
| Hordeivirus | 3 | Separate | TGB | 22K | t-RNATyr | seed |
| Pecluvirus | 2 | RT | TGB | 23K | t-RNAVal | plasmidophorid + seed |
| Pomovirus | 3 | RT | TGB | 20K+RT | t-RNAVal | plasmodiophorid |
| Tobamovirus | 1 | RT | “30K” | 17–18K | t-RNAHis | mechanical |
| Tobravirus | 2 | RT | “30K” | 22–24K | t-RNA- | nematode |
a Relation of the RdRP encoding ORF to the replication protein (methyltransferase, helicase) encoding ORF; RT, in a 3′-proximal region expressed by ribosomal readthrough.
b MP, Movement protein either of the “30K” superfamily or a triple gene block (TGB).
c CP, Coat protein size in kDa (with indication of a readthrough domain (RT) at the C-terminus if present).
d t-RNAVal/Tyr/His/?/-, t-RNA like structure accepting valine, tyrosine, histidine, unknown amino acid, or not aminoacylated respectively.
Relationships within the family
Phylogenetic trees obtained using the entire replication protein including the RdRP (or the conserved Mtr, Hel and RdRP domains) correlate well with genome organisation to support the existence of the different genera. However, these genera are usually reliably delineated, regardless of the protein used (Figures 1. Virgaviridae - 5. Virgaviridae). Using both the replication protein and the coat protein, the genus Tobamovirus separates substantially from the remaining genera. Furovirus and Pomovirus occur together on the same branch in all trees as do Pecluvirus, Goravirus and Hordeivirus.
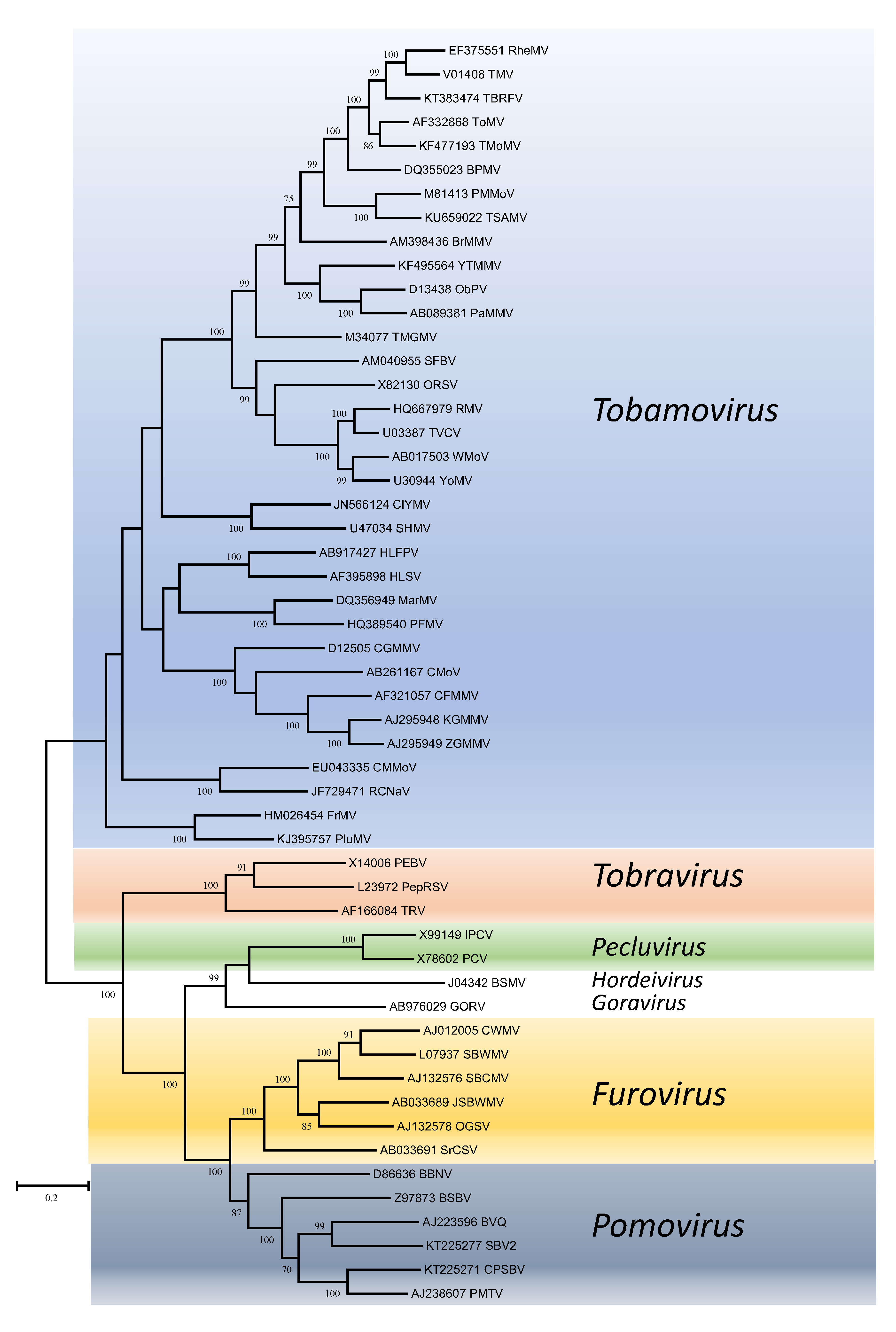 |
| Figure 1. . Virgaviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the replication proteins (including Mtr and Hel domains) of viruses in the family Virgaviridae. The analyses were conducted in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-135263.7399) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Virus abbreviations are explained in the tables of member species and other viruses. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
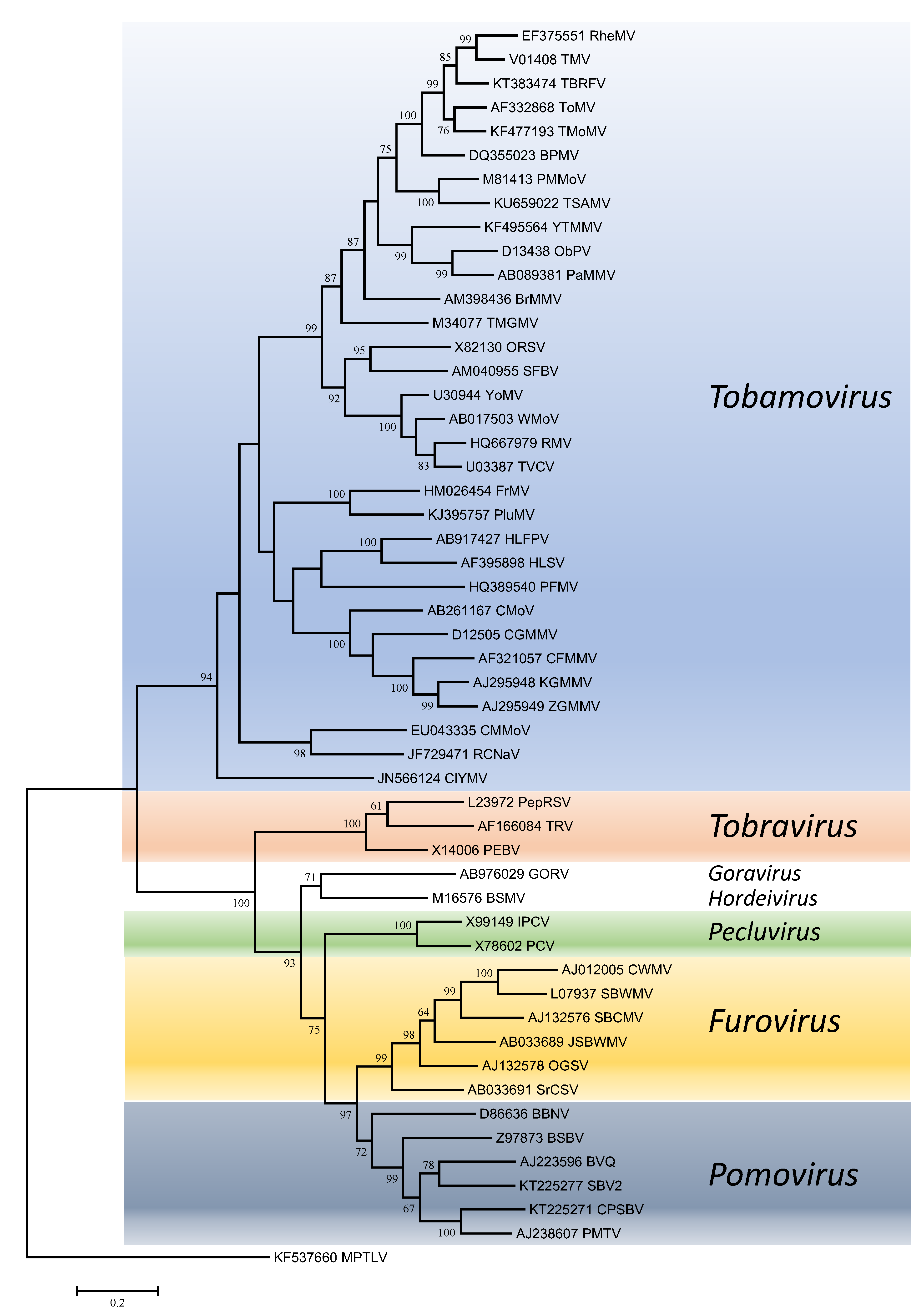 |
| Figure 2. . Virgaviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the RdRP domain of viruses in the family Virgaviridae. The analyses were conducted in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-57431.3254) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Virus abbreviations are explained in the tables of member species and other viruses. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
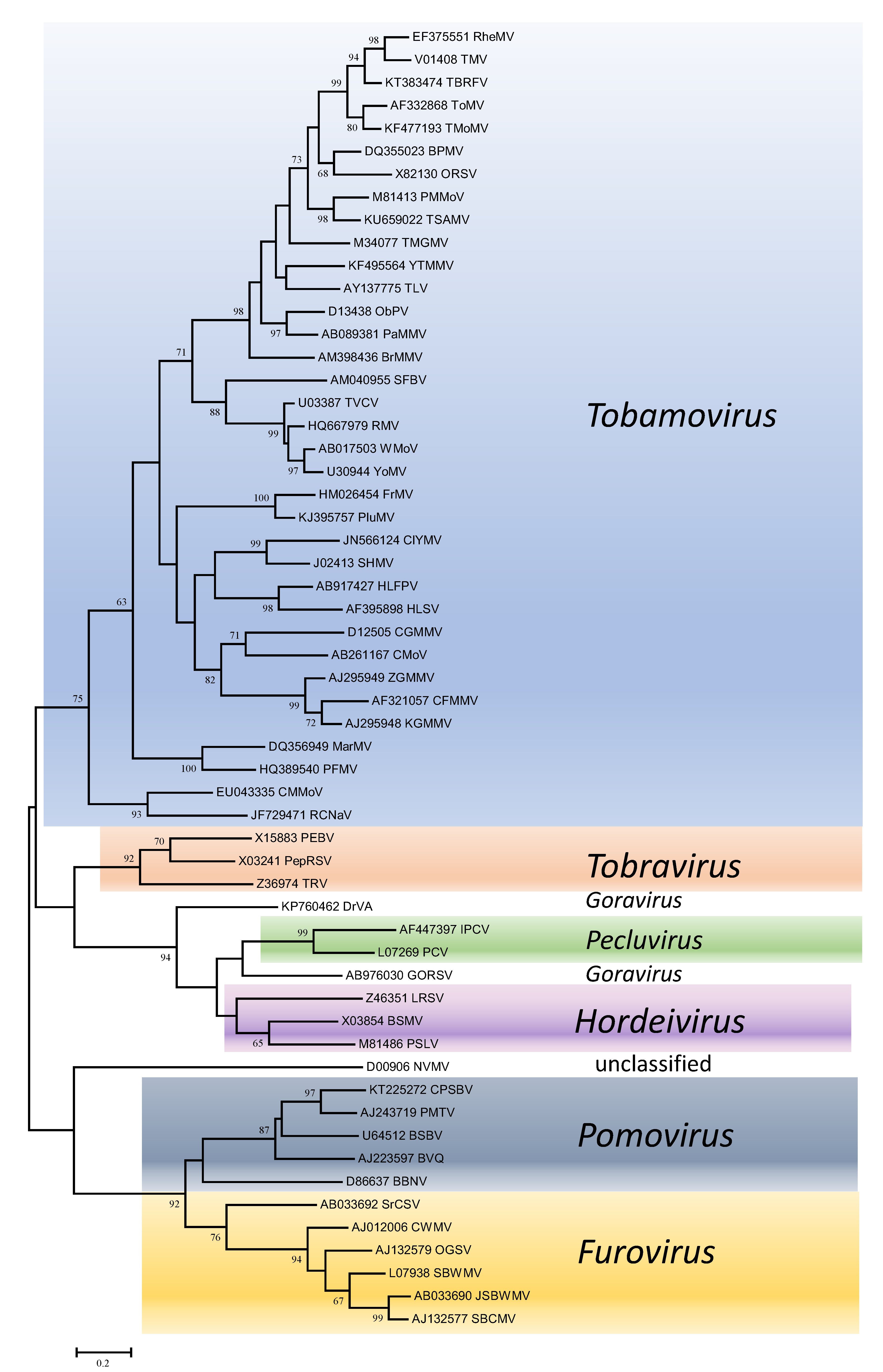 |
| Figure 3. . Virgaviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the coat proteins of viruses in the family Virgaviridae. The analyses were conducted in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-19333.2064) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Virus abbreviations are explained in the tables of member species and other viruses. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
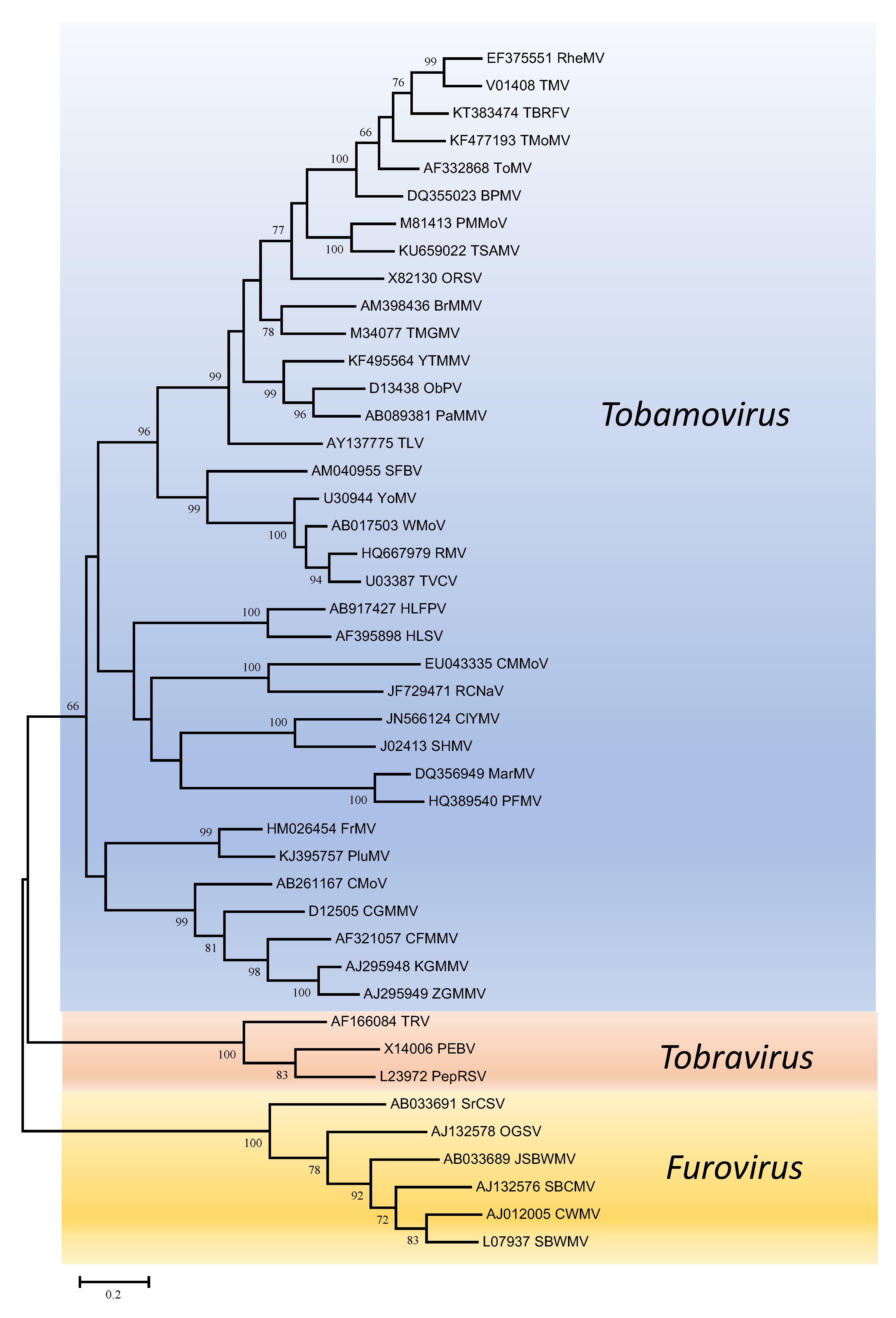 |
| Figure 4. . Virgaviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the movement proteins of viruses in the family Virgaviridae. Only viruses with a 30K-like movement protein were included in the analyses. The analyses were conducted in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-24269.0075) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Virus abbreviations are explained in the tables of member species and other viruses. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
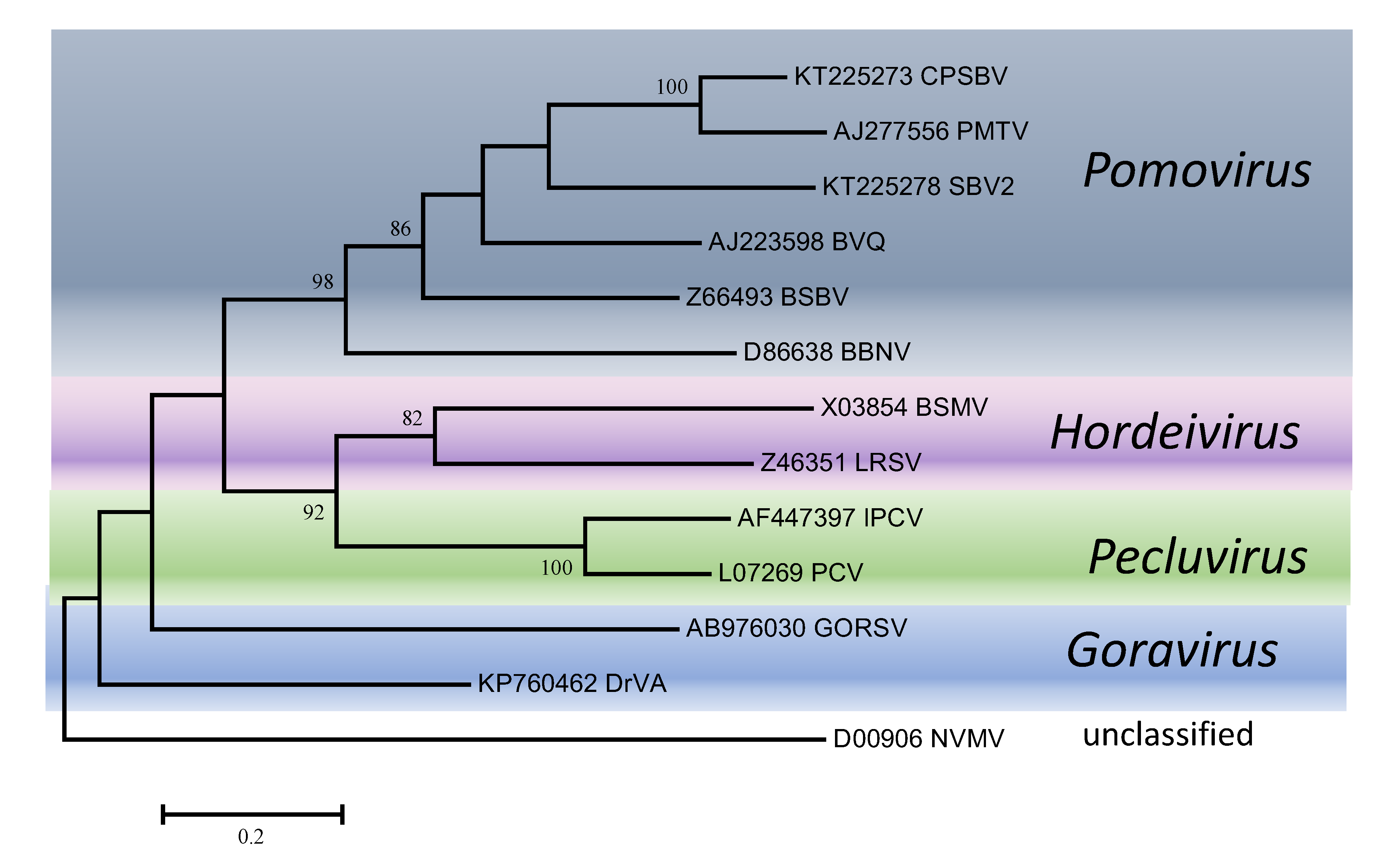 |
| Figure 5. . Virgaviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the first triple gene block protein (TGB1) of viruses in the family Virgaviridae. The analyses were conducted in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-13286.8458) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Virus abbreviations are explained in the tables of member species and other viruses. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Within genera, only members of the genus Tobamovirus have enough diversity for particular subgroupings to be distinguished. Here, there are clearly groupings of closely-related viruses infecting similar hosts. The most obvious are those infecting cucurbits (cucumber fruit mottle mosaic virus, cucumber green mottle mosaic virus, cucumber mottle virus, kyuri green mottle mosaic virus, zucchini green mottle mosaic virus), cruciferous plants (ribgrass mosaic virus, turnip vein-clearing virus, wasabi mottle virus, youcai mosaic virus) and solanaceous plants (brugmansia mild mottle virus, obuda pepper virus, paprika mild mottle virus, pepper mild mottle virus, rehmannia mosaic virus, tobacco mild green mosaic virus, tobacco mosaic virus, tomato brown rugose fruit virus, tomato mosaic virus, tropical soda apple mosaic virus, yellow tailflower mild mottle virus).
Relationships with other taxa
The replication proteins are related to those of other viruses with alpha-like replicases, and more particularly to those in the families Closteroviridae and Bromoviridae and the unassigned genera Idaeovirus, Blunervirus, Cilevirus and Higrevirus (Figure 6. Virgaviridae). The only other viruses with rod-shaped virions are those classified in the genus Benyvirus (family Benyviridae) and they are distinguished because they are distantly related in phylogenetic analyses and because (unlike the members of the Virgaviridae) they have a polyadenylated genome and a polymerase that is processed by autocatalytic protease activity.
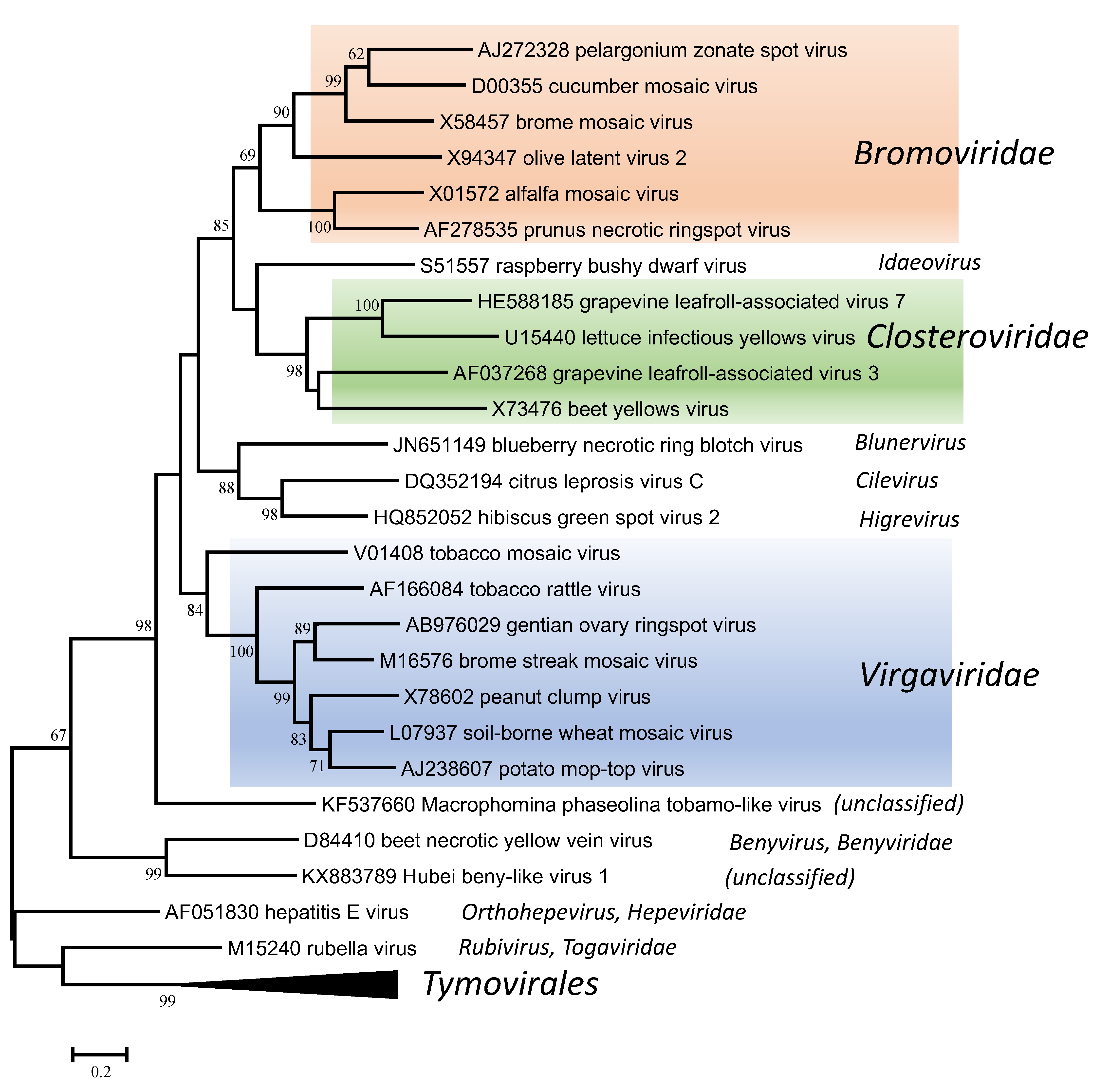 |
| Figure 6. . Virgaviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the RdRP proteins of viruses in the family Virgaviridae and some other viruses. The analyses were conducted in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-42095.8020) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Representative isolates of the type species of the genera Benyvirus, Blunervirus, Cilevirus, Higrevirus, Idaeovirus, Orthohepevirus and Rubivirus and of each of the genera in the families Bromoviridae, Closteroviridae and Virgaviridae and the order Tymovirales were used in the analysis. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Nicotiana velutina mosaic virus | D00906* | NVMV |
Virus names and virus abbreviations are not official ICTV designations.
* partial genome

