Family: Potyviridae
Genus: Ipomovirus
Distinguishing features
Ipomoviruses are distinguished from members of other genera by their mode of transmission being by whiteflies, and by their separate branching on phylogenetic analyses.
Virion
Morphology
Virions are flexuous filaments 800–950 nm long.
Physicochemical and physical properties
Virion Sedimentation coefficient S20,w is 155S for sweet potato mild mottle virus (SPMMV).
Nucleic acid
Virions contain a single molecule of linear, positive-sense ssRNA of about 9.7 kb; virions contain 5% RNA by weight.
Proteins
The viral CP is a single polypeptide of 302–378 aa (35–41 kDa).
Genome organization and replication
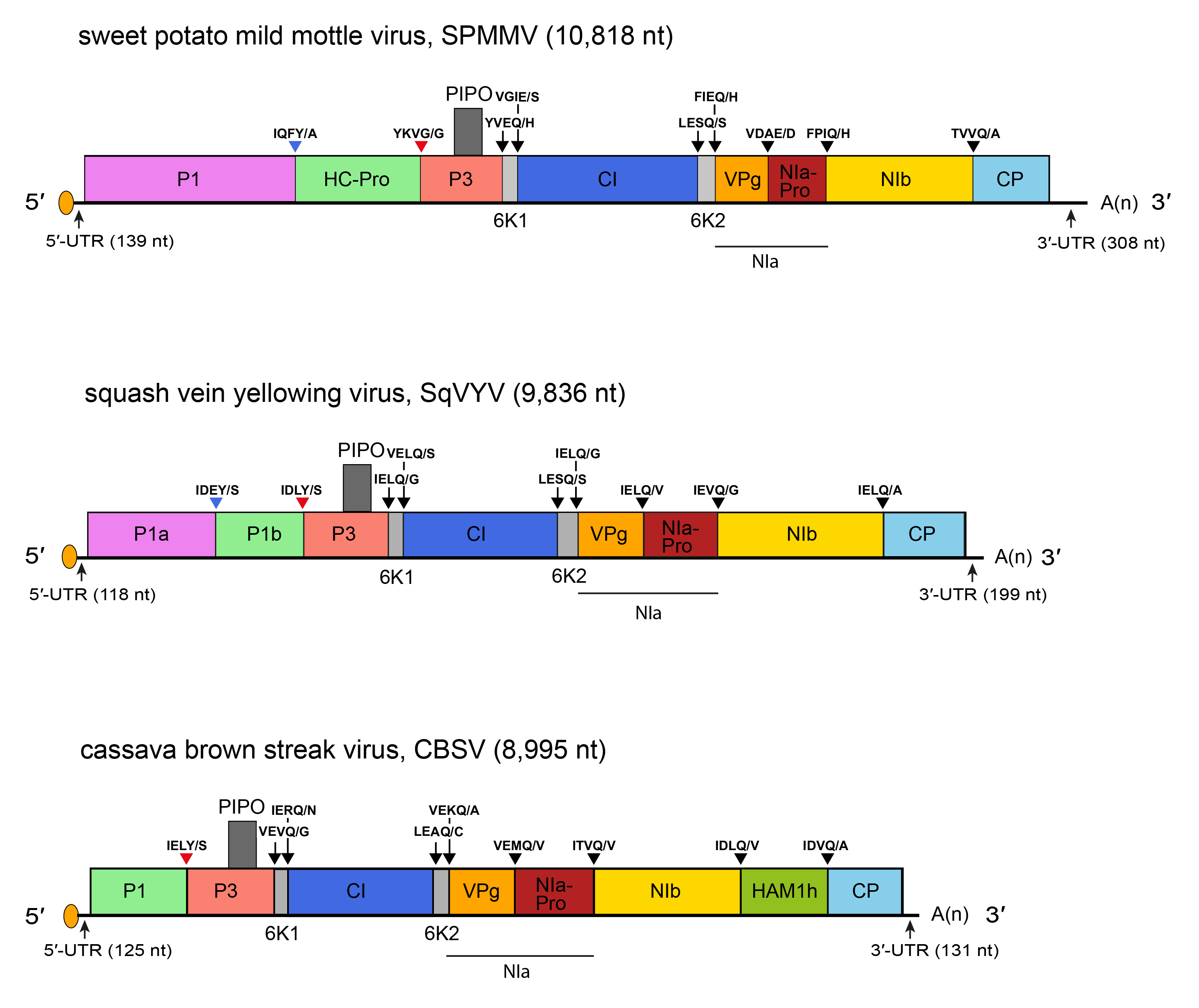
Ipomoviruses exhibit unusual structural variability (Figure 1.Ipomovirus). The structure and organization of the SPMMV genome is similar to that of members of the genus Potyvirus (Figure 2.Potyviridae), but some motifs of HC-Pro and CP characteristic of members of the genus Potyvirus are incomplete or missing, which may account for its vector relations. The unusually large P1 protein (83 kDa) of SPMMV contains no obvious AlkB domain and hence differs from that found in members of the genus Brambyvirus. Coccinia mottle virus (CocMoV), cucumber vein yellowing virus (CVYV), and squash vein yellowing virus (SqVYV) differ from SPMMV and tomato mild mottle virus (TMMoV) by encoding two P1-like serine proteases (P1a and P1b) but no HC-Pro (Li et al., 2008). P1b functions as a suppressor of RNA silencing (Valli et al., 2006). Cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV) differ from SPMMV by having no HC-Pro and differ from CVYV and SqVYV by having only P1b, which suppresses silencing. Additionally, CBSV and UCBSV contain a Maf/HAM1-like sequence recombined into the NIb/CP junction, accommodating heterologous genes in engineered infectious potyvirus clones (Mbanzibwa et al., 2009, Mbanzibwa et al., 2011). Homology of HAM1h with cellular Maf/HAM1 NTP pyrophosphatases suggests that HAMh1 might intercept non-canonical NTPs to reduce mutation rates of viral RNA.
|
|
| Figure 1.Ipomovirus. Schematic diagrams of the ipomoviruses sweet potato mild mottle virus (SPMMV), squash vein yellowing virus (SqVYV) and cassava brown streak virus (CBSV) genomes. The ssRNA genome is represented by a line and the polyprotein ORF by an open box. Conventions are as for the potyvirus genome organization map (Figure 2.Potyviridae). Activities of most mature proteins are postulated by analogy with genus Potyvirus. CVYV, CocMoV and SqVYV contain two P1-like serine proteases (P1a and P1b), of which P1b functions as a suppressor of RNA silencing. CBSV also contains P1b, which suppresses silencing and, additionally, carries a Maf/HAM1-like sequence recombined into the NIb/CP junction. HAMh1 might intercept non-canonical NTPs to reduce mutation rates of viral RNA. Not to scale. |
Biology
Host range
The natural host range of SPMMV is wide, with more than nine plant families susceptible, whereas the host range of CBSV, UCBSV, TMMoV, CVYV, CocMoV and SqVYV is less known apart from the hosts which they have been found to infect in the field (Winter et al., 2010).
Transmission
CBSV, UCBSV, TMMoV, CVYV and SqVYV are transmitted by the whitefly Bemisia tabaci in a non-persistent manner. B. tabaci may also be the vector of SPMMV, but this is not fully confirmed. All ipomoviruses are transmissible experimentally by mechanical inoculation and by grafting
Antigenicity
Moderately immunogenic. No serological relationships with other members of the family Potyviridae have been found.
Species demarcation criteria
See discussion under family description.