Family: Potyviridae
Genus: Bevemovirus
Distinguishing features
The genus includes a single species, Bellflower veinal mottle virus; members of this species, like macluraviruses and arepaviruses, lack the P1 region present in most potyvirids. The HC-Pro lacks the conserved potyvirus aphid transmission motifs R/KITC and PTK. Isolates of bellflower veinal mottle virus share low (22–27%) nucleotide and amino acid sequence identities to macluraviruses.
Virion
Nucleic acid
Bellflower veinal mottle virus has a positive-sense RNA genome of 8,259 nt.
Genome organization and replication
Genome organization (Figure 1.Bevemovirus) resembles that of members of the genus Macluravirus (Figure 1.Macluravirus).
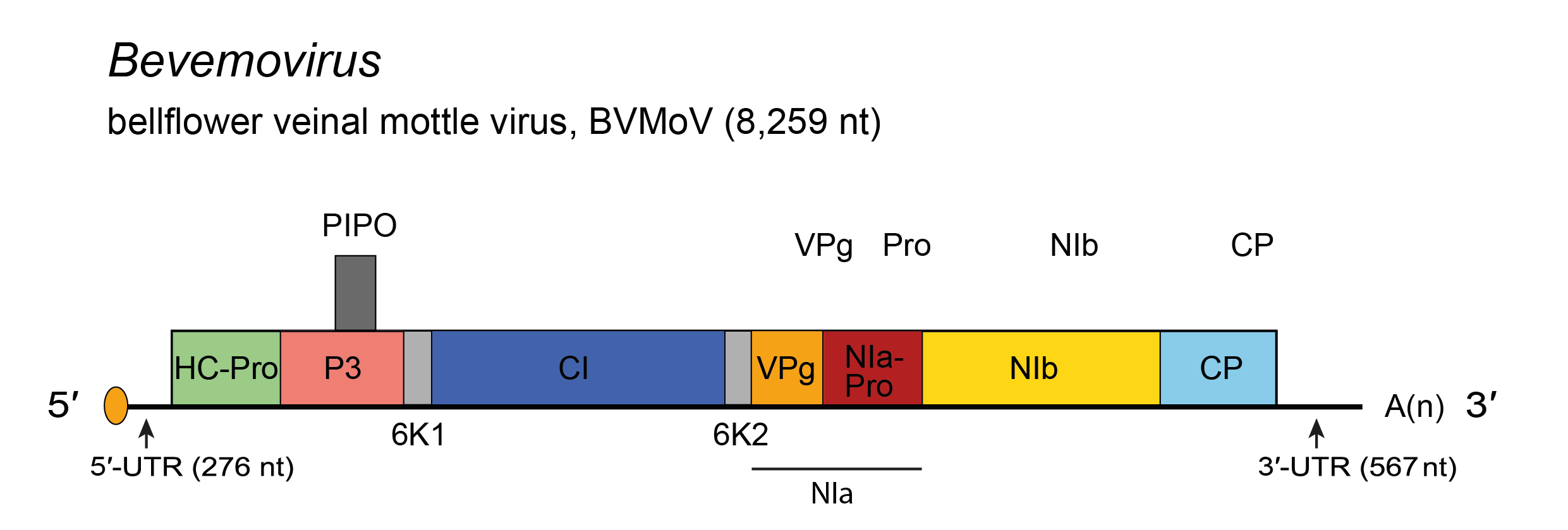 |
| Figure 1.Bevemovirus. Schematic diagram of the bellflower veinal mottle virus (BVMoV) genome. The polyprotein ORF is indicated by the large open box divided into putative mature proteins. The pretty interesting Potyviridae protein (PIPO) is represented by a small box. The untranslated regions (UTR) are represented by lines on each end of the large ORF. Activities of mature proteins are postulated by analogy with genus Potyvirus. Conventions are as for the potyvirus genome organization map (Figure 2.Potyviridae). BVMoV lacks a P1 cistron. Not to scale. |
Biology
Isolates of Bellflower veinal mottle virus were obtained from bellflower (Campanula takesimana) that exhibited veinal mottle symptoms in South Korea (Seo et al., 2017). The virus is probably not aphid transmitted because the HC-Pro lacks the conserved potyvirus aphid transmission motifs R/KITC and PTK.
Species demarcation criteria
See discussion under family description.

