Family: Potyviridae
Genus: Arepavirus
Distinguishing features
Arepaviruses resemble members of the genus Macluravirus. Their genomes differ from other potyvirids in that they encode two proteinases in two HC-Pro-like proteins (HC-Pro1 and HC-Pro2), at the 5′-end of the genome, but no P1 cistron (Figure 1.Arepavirus). Both HC-Pro-like proteins encode conserved cysteine protease domains of the peptidase C6 superfamily. The Gly, Cys and His residues, corresponding to the catalytic activity site, are present in both HC-Pro1 and HC-Pro2. The genomes of arepaviruses also differ from those of macluraviruses in that they are larger (about 9.4 kb vs 8 kb) and with pairwise identities between members of different genera of <46%.
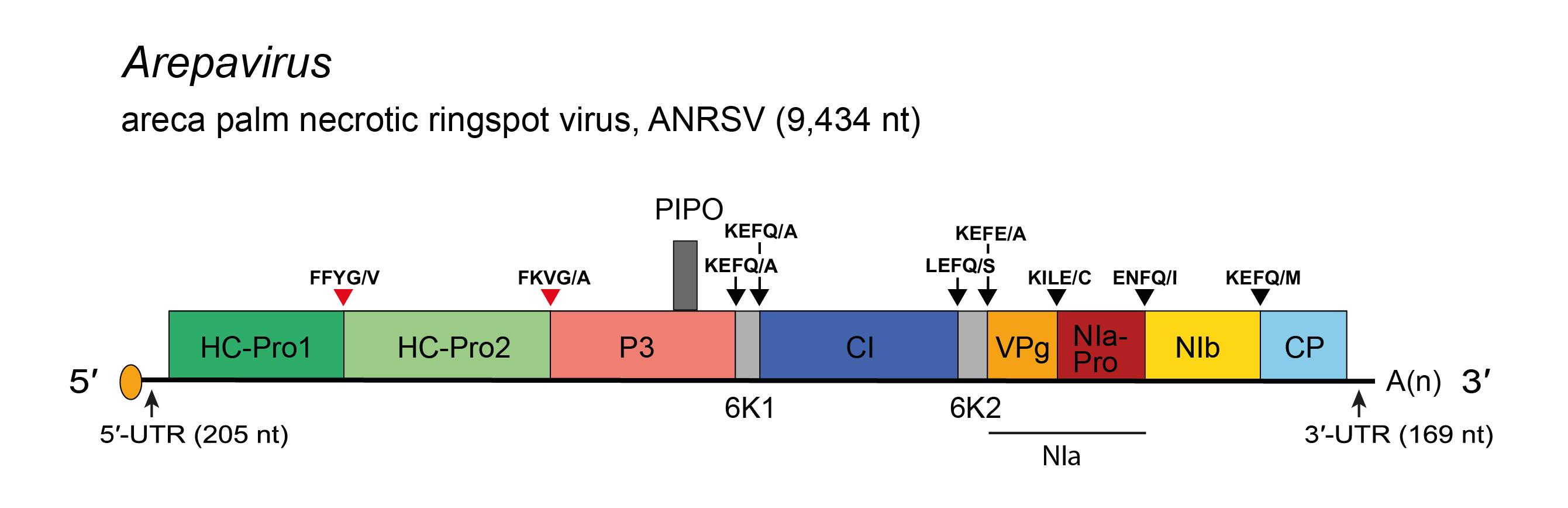 |
| Figure 1.Arepavirus. Schematic diagram of areca palm necrotic ringspot virus (ANRSV) genome. The polyprotein ORF is indicated by the large open box divided into putative mature proteins. The pretty interesting Potyviridae protein (PIPO) is represented by a small box. The untranslated regions (UTR) are represented by lines on each end of the large ORF. Activities of mature proteins are postulated by analogy with genus Potyvirus. Conventions are as for the potyvirus genome organization map (Figure 2.Potyviridae). ANRSV lacks a P1 cistron and has two cysteine proteinases in tandem in an HC-Pro-like cistron that is presumed to be processed into two mature proteins, named HC-Pro1 and HC-Pro2. Not to scale. |
Virion
Morphology
Flexuous filamentous potyvirid-like particles of about 15×780 nm.
Nucleic acid
Virions contain one molecule of linear positive-sense ssRNA of about 9.4 kb.
Proteins
Arepaviruses have a single coat (capsid) protein (CP) species of 33–34 kDa.
Genome organization and replication
The complete genome sequences of two arepaviruses are available: areca palm necrotic spindle-spot virus and areca palm necrotic ringspot virus. The genomes are ~9.4 kb, encoding a polyprotein of 3,019 amino acids. The genome organization and the encoded protein products resemble those of macluraviruses. Like macluraviruses, arepaviruses lack a cistron that encodes a P1 protein, but unlike them, arepaviruses encode nine proteinase cleavage sites, not eight. Uniquely, there are two cysteine proteinases in tandem in the HC-Pro-like cistron, and putative cleavage sites exist for each. Therefore, the HC-Pro is assumed to be processed into two mature proteins, named HC-Pro1 and HC-Pro2. The typical potyviral PIPO ORF is located within P3; it is predicted to be translated by frameshift after polymerase slippage at GA7. The ten mature proteins (listed from N-terminus) are denoted HC-Pro1, HC-Pro2, P3, 6K1, CI, 6K2, VPg, NIa-Pro, NIb and CP. Other conserved potyvirid motifs were detected. Other than lacking a P1 cistron and having two HC-Pro proteins, the arepavirus genome organization and replication strategy appear typical of other potyvirids (Figure 1.Arepavirus).
Biology
Host range
Both known arepaviruses infect plants of Areca catechu (Areca palm) on Hainan Island, China (Yang et al., 2018, Yang et al., 2019).
Transmission
Unknown, but believed to be vectored by insects. The HC-Pro motifs PTK and KITC and the CP motif DAG, which are involved in aphid transmission in some other potyvirids, are absent, suggesting that aphids are not vectors of these viruses.
Species demarcation criteria
See discussion under family description.

