Family: Potyviridae
Genus: Celavirus
Distinguishing features
The isolate of the single celavirus described to date, celery latent virus (CeLV), was distinguished by a very short 5′-UTR of 13 nt and an unusually long 3′-UTR of 586 nt, which is not polyadenylated. The genome is 11.5 kb in size. Typical potyvirid-like protease cleavage sites were not present in the N-terminal two-thirds of the polyprotein, although this region possesses a number of sequence motifs typical of potyvirids. A clearly definable HC-Pro cistron was lacking; characteristic potyvirid motifs of the HC-Pro were absent. The 6K peptides typical of most potyvirids were not identified. A signal peptide of 23 aa was found at the N-terminal part of P1 protein, likely guiding proteins to or into the endoplasmic reticulum. The virus was not transmitted by five tested aphid species.
Virion
Morphology
Virions are flexuous filaments 885–900 nm long.
Nucleic acid
Virions contain a single molecule of linear, positive-sense ssRNA of about 11.5 kb.
Proteins
The viral CP is a single polypeptide of 380 aa (41.8 kDa).
Genome organization and replication
The genome contains a single large open reading frame (ORF) that is flanked by an unusually-short 5′-untranslated region (UTR) of 13 nt and an unusually-long 3′-UTR of 586 nt (Figure 1.Celavirus). Unique among potyvirids, the 3′-UTR is not polyadenylated. The large ORF is deduced to encode a polyprotein of 3640 amino acids. Like other potyvirids, the CeLV polyprotein has a putative P3N-PIPO ORF, but the PIPO shares very low amino acid sequence identities (about 18 %) with those of other potyvirids. Typical potyvirid-like protease cleavage sites were not identified for the N-terminal two-thirds of the polyprotein, although this region possesses a number of sequence motifs typical of potyvirids. For example, the PIPO ORF is present, as are CI and NIb motifs. However, many other characteristic potyvirid motifs were not present, including motifs characteristic of the HC-Pro such as KITC, IGN, CC/SC, FRNK, PTK and GYCY. No cytoplasmic cylindrical (“pinwheel”) inclusions were visible in infected tissue.
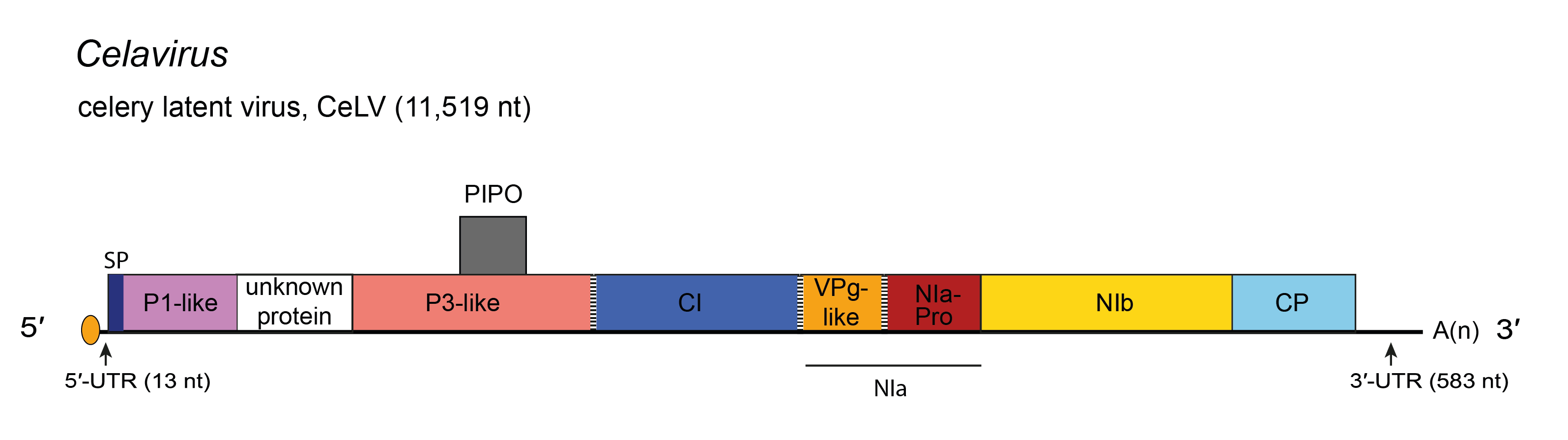 |
| Figure 1.Celavirus. Schematic diagram of the celavirus celery latent virus (CeLV) genome. The polyprotein ORF is indicated by the large open box divided into putative mature proteins. The pretty interesting Potyviridae protein (PIPO) is represented by a small box. The untranslated regions (UTR) are represented by lines on each end of the large ORF. Activities of mature proteins are postulated by analogy with genus Potyvirus. Conventions are as for the potyvirus genome organization map (Figure 2.Potyviridae). SP, Signal Peptide. Not to scale. |
Biology
Host range
Celery latent virus (CeLV) was first described in the 1960s and 1970s (Brandes and Luisoni 1966, Bos et al., 1978) from the Netherlands in asymptomatic celeriac (Apium graveolens var. rapaceum). Sixteen experimental hosts have been described, including species of Amaranthus, Apium, Chenopodium, Nicotiana, Pisum, Spinacia and Trifolium (Bos et al., 1978). These authors were unable to transmit CeLV using five aphid species, but seed transmission was high in celeriac (34%) and C. quinoa (67%), and confirmed in Amaranthus.
Further experiments have repeated and confirmed much of the earlier work on CeLV (Rose et al., 2019). Isolate CeLV-Ag097 was described from an asymptomatic plant of leaf celery (A. graveolens var. secalinum) in Italy. When inoculated to other apiaceous plants, it asymptomatically infected plants of celeriac and celery (A. graveolens var. dulce). Systemic infection occurred in Chenopodium quinoa, where symptoms consisted of yellow (ring-) spots, and symptomless systemic infection on N. benthamiana was recorded.
Transmission
CeLV-Ag097 was sap- and seed-transmissible, and it was not transmissible by five aphid species.
Species demarcation criteria
See discussion under family description.

