Family: Crepuscuviridae
Jens H. Kuhn, Nolwenn M. Dheilly, Sandra Junglen, Sofia Paraskevopoulou (Σοφία Παρασκευοπούλου), Mang Shi (施莽) and Nicholas Di Paola
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Nicholas Di Paola ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2023
Summary
Crepuscuviridae is a family for negative-sense RNA viruses with genomes of about 12.0 kb (Table 1.Crepuscuviridae). The family includes a single genus with one species for a virus found in insects in Carabobo State, Venezuala. The crepuscuvirid genome contains open reading frames (ORFs) that encode a glycoprotein (GP), a nucleoprotein (NP), and a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain.
Table 1.Crepuscuviridae. Characteristics of members of the family Crepuscuviridae
| Characteristic | Description |
| Example | megalopteran chu-related virus 119 (MW039261), species Aqualaruvirus sialis, genus Aqualaruvirus |
| Virion | Unknown |
| Genome | About 12.0 kb of nonsegmented negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Megalopteran insects |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Monjiviricetes, order Jingchuvirales; the family includes one genus and one species |
Virion
Nucleic acid
Crepuscuvirids have nonsegmented linear negative-sense RNA genomes with total lengths of about 12.0 kb (Käfer et al., 2019).
Genome organization and replication
Crepuscuvirid genomes have three ORFs that encode a GP, an NP, and an L protein (Käfer et al., 2019) (Figure 1.Crepuscuviridae). The replication cycle of crepuscuvirids remains to be elucidated.
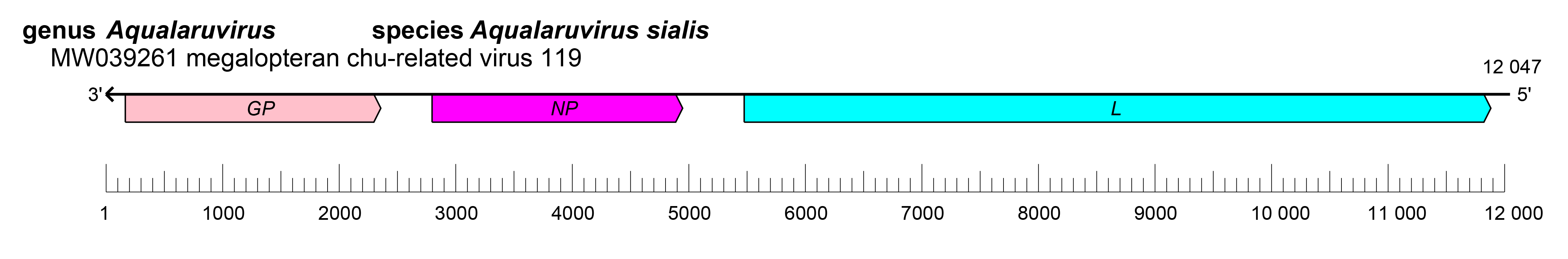 |
| Figure 1.Crepuscuviridae. Genome organization of megalopteran chu-related virus 119. ORFs are colored according to the predicted protein function. GP, glycoprotein gene; NP, nucleoprotein gene; L, large protein gene. |
Biology
The only classified crepuscuvirid, megalopteran chu-related virus 119 (MCrV119), has been associated with megalopteran insects (Corydalidae) sampled in Carabobo State, Venezuela (Käfer et al., 2019).
Derivation of names
Aqualaruvirus: from the Latin aqua, meaning “water” and the Latin larua, meaning “ghost, evil spirit, demon”
Crepuscuviridae: from Latin crepusculum, meaning “twilight, dim”
sialis: erroneously derived from the fly family Sialidae, a sister family to Corydalinae in the order Megaloptera. The virus megalopteran chu-related virus OKIAV119 (MCrV-119) was discovered by HTS in a corydalid fly sampled in Bejuma, Carabobo State (Estado Carabobo), Venezuela
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Not applicable (the only genus includes one species).
Relationships within the family
Only one virus has been described in the family.
Relationships with other taxa
Viruses in the family Crepuscuviridae are most closely related to jingchuviral aliusvirids, chuvirids, myriavirids, and natarevirids (Käfer et al., 2019, Di Paola et al., 2022) (Figure 2.Crepuscuviridae).
 |
| Figure 2.Crepuscuviridae. Phylogenetic relationships of crepuscuvirids. Maximum-likelihood tree (midpoint-rooted) inferred by using large protein gene (L) sequences. Sequences were initially aligned by MAFFT version 7 (https://mafft.cbrc.jp/alignment/software/) in Geneious version R9 (http://www.geneious.com) and realigned using ClustalW (https://www.genome.jp/tools-bin/clustalw). The tree was estimated using PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/ , a subtree pruning and regrafting (SPR) topology searching algorithm, and a Bayes branch support algorithm. Numbers near nodes on the trees indicate bootstrap values as percentages. Tree branches are scaled to nucleotide substitutions per site (scale bar). Two mononegavirals were included as an outgroup. |

