Family: Aliusviridae
Jens H. Kuhn, Nolwenn M. Dheilly, Sandra Junglen, Sofia Paraskevopoulou (Σοφία Παρασκευοπούλου), Mang Shi (施莽), and Nicholas Di Paola
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Nicholas Di Paola, [email protected]
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2023
Summary
Aliusviridae is a family of negative-sense RNA viruses with genomes of 9.9–15.3 kb (Table 1.Aliusviridae). These viruses have been found in insects in Asia, Australia, Europe, North America, and South America. The family includes two genera with 12 species for 12 viruses. The aliusvirid genome contains two to four open reading frames (ORFs) that encode a glycoprotein (GP), nucleoprotein (NP), a large (L) protein containing RNA-directed RNA polymerase (RdRP) domain, and/or proteins of unknown function.
Table 1.Aliusviridae Characteristics of members of the family Aliusviridae
| Characteristic | Description |
| Example | Culverton virus (MN167499), species Ollusvirus culvertonense, genus Ollusvirus |
| Virion | Unknown |
| Genome | 9.9–15.3 kb of nonsegmented negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Coleopteran, dipteran, hemipteran, hymenopteran, and siphonapteran insects |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Monjiviricetes, order Jingchuvirales; the family includes two genera and 12 species |
Virion
Morphology
Unknown.
Nucleic acid
Aliusvirids have nonsegmented linear negative-sense RNA genomes with total lengths of 9.9–15.3 kb (Li et al., 2015, Schoonvaere et al., 2016, Shi et al., 2016, Harvey et al., 2019, Käfer et al., 2019, Wang et al., 2019, Han et al., 2020, Wu et al., 2020, Chang et al., 2021).
Genome organization and replication
Aliusvirid genomes have two to four ORFs that encode a GP, NP, and L protein, and/or proteins of unknown function (Li et al., 2015, Schoonvaere et al., 2016, Shi et al., 2016, Harvey et al., 2019, Käfer et al., 2019, Wang et al., 2019, Han et al., 2020, Wu et al., 2020, Chang et al., 2021).
The replication cycle of aliusvirids remains to be elucidated.
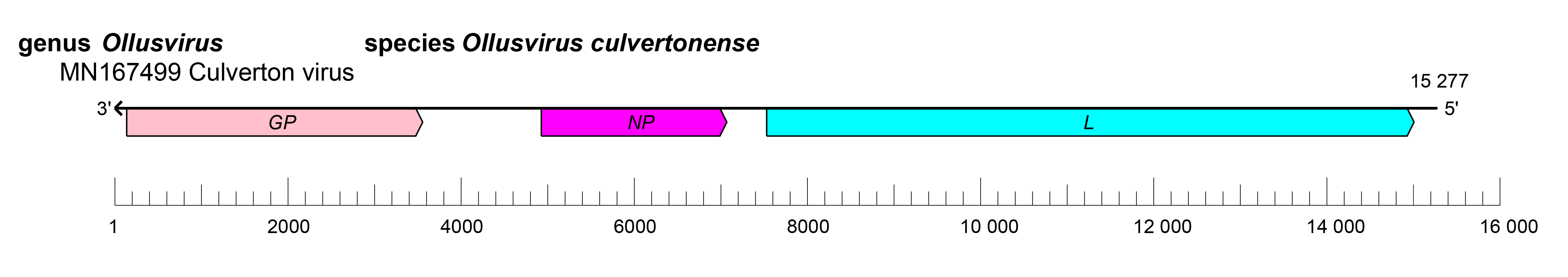 |
| Figure 1.Aliusviridae. Genome length, organisation, and position of open reading frames (ORFs) of a representative virus in family Aliusviridae, Culverton virus (GenBank MN167499). ORFs are indicated as boxes, coloured according to the predicted protein function. GP, glycoprotein gene; NP, nucleoprotein gene; L, large protein gene. |
Biology
Aliusvirids have been associated with insects.
Derivation of names
Aliusviridae: from Latin alius, meaning “other, another”
coleopteri: from Húběi coleoptera virus 3
culvertonense: from Culverton virus
hanchengense: from Hánchéng leafhopper mivirus
hymenopteri: from hymenopteran chu-related virus 123
insectii: from insect
nomadae: from Nomada lathburiana mononega-like virus
Obscuruvirus: from Latin obscurus, meaning “dark, dusky, shadowy”
Ollusvirus: from Estonian ollus, meaning “substance, matter, material
oropsyllae: from Oropsylla silantiewi mononega-like virus 2
rhagoveliae: from Rhagovelia obesa mononega-like virus
scaldisense: from Scaldis River bee virus
shayangense: from Shāyáng fly virus 1
taiyuanense: from Tàiyuán leafhopper virus
Genus demarcation criteria
Members of different aliusvirid genera have L protein sequences with <31% amino-acid identity (Di Paola et al., 2022).
Species demarcation criteria
Members of different aliusvirid species have L protein sequences with <90% amino-acid identity (Di Paola et al., 2022).
Relationships within the family
Phylogenetic relationships of members of the family Aliusviridae are shown in Figure 2.Aliusviridae.
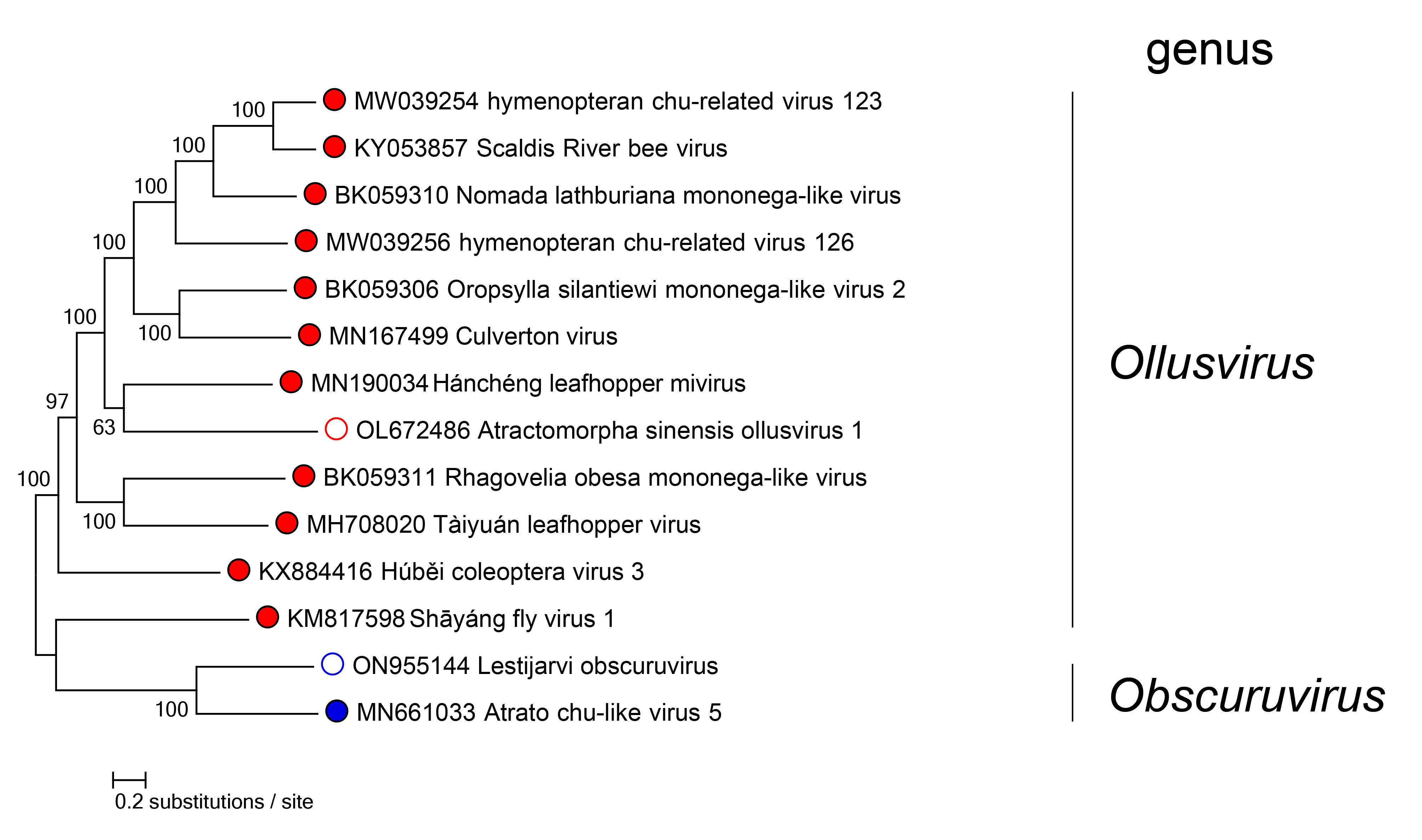 |
| Figure 2.Aliusviridae. Phylogenetic relationships of aliusvirids. Maximum-likelihood tree (midpoint-rooted) inferred by large protein gene (L) nucleotide sequences. Sequences were initially aligned by MAFFT version 7 (https://mafft.cbrc.jp/alignment/software/) in Geneious version R9 (http://www.geneious.com) and realigned using ClustalW (https://www.genome.jp/tools-bin/clustalw). Trees were inferred in FastTree version 2.1 (http://www.microbesonline.org/fasttree/) using a General Time Reversible (GTR) model with 20 Gamma-rate categories, 5,000 bootstrap replicates, and exhaustive search parameters (-slow) and pseudocounts (-pseudo). Numbers near nodes on the trees indicate bootstrap values as percentages. Tree branches are scaled to nucleotide substitutions per site (scale bar). |
Relationships with other taxa
Aliusvirids are most closely related to jingchuviral chuvirids, crepuscuvirids, myriavirids, and natarevirids (Di Paola et al., 2022).

