Subfamily: Mammantavirinae
Genus: Thottimvirus
Distinguishing features
Imjin virus (MJNV) and Thottapalayam virus (TPMV) are the only classified thottimviruses. Thottimviruses infect eulipotyphla (Carey et al., 1971, Song et al., 2009).
Virion
Virions are unknown.
Nucleic acid
Thottimviruses have tri-segmented negative-sense RNA genomes of 11.7–12.0 kb (small [S] segment: 1.5–1.6 kb; medium [M] segment: 3.6 kb; large [L] segment: 6.6 kb) (Carey et al., 1971, Song et al., 2009).
Proteins
Based on sequence data only, thottimviruses likely express three structural proteins: nucleoprotein (N), glycoprotein precursor (GPC), and large protein (L) (Carey et al., 1971, Song et al., 2009, Kang et al., 2014, Kang et al., 2016).
Genome organization and replication
The S segment encodes N, the M segment encodes GPC, and the L segment encodes L (Figure 1 Thottimvirus). Thottimvirus genomic segments are expected to assume circular forms via non-covalent binding of complementary and conserved 3′- and 5′-terminal sequences.
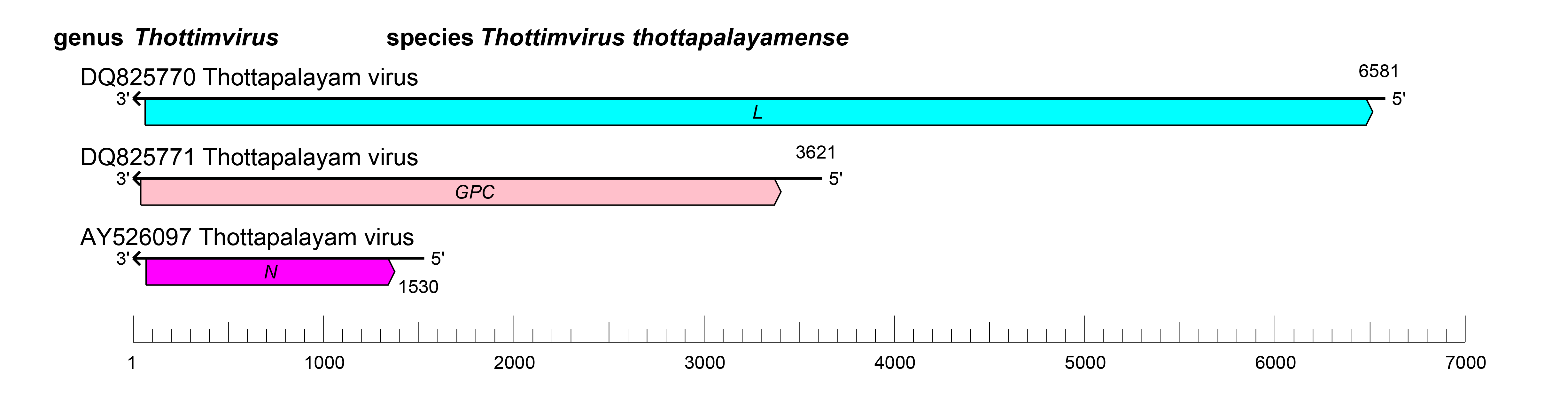 |
| Figure 1 Thottimvirus. Schematic representation of thottimvirus genome organization. The 5′- and 3′-ends of each segment (S, M and L) are, by analogy to other hantavirids, predicted to be complementary at their termini, likely promoting the formation of circular ribonucleoprotein complexes within the virion. |
Biology
MJNV and TPMV infects Ussuri white-toothed shews (soricid Crocidura lasiura (Dobson, 1890)) in China and South Korea and Asian house shrews (soricid Suncus murinus (Linnaeus, 1766)) in China, India, and Nepal, respectively (Carey et al., 1971, Song et al., 2009, Guo et al., 2011, Kang et al., 2011b, Lin et al., 2014, Sun et al., 2017). Unclassified, potential thottimviruses infect talpid moles in China and soricid shrews in Tanzania (Kang et al., 2014, Kang et al., 2016). TPMV has been isolated (Carey et al., 1971), but thottimvirus biology remains to be elucidated.
Species demarcation criteria
Demarcation of species is based upon DivErsity pArtitioning by hieRarchical Clustering (DEmARC) analysis) using concatenated deduced S, M, and L segment expression product sequences (Laenen et al., 2019).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Dàhónggōu Creek virus | L: HQ616595* | DHCV | (Kang et al., 2016) |
| Kilimanjaro virus | S: JX193698; M: JX193699*; L: JX193700 | KMJV | (Kang et al., 2014) |
| Uluguru virus | S: JX193695; M: JX193696*; L: JX193697 | ULUV | (Kang et al., 2014) |
Virus names and virus abbreviations are not official ICTV designations.
*Coding region sequence incomplete.

