Subfamily: Mammantavirinae
Genus: Orthohantavirus
Distinguishing features
Sixty different viruses are classified in the genus Orthohantavius. These viruses typically infect specific soricid and talpid eulipotyphla and muroid and possibly dipodoid rodents. Orthohantaviruses are the only hantavirids known to cause disease in humans (Schmaljohn and Hjelle 1997, Jonsson et al., 2010, Kuhn and Schmaljohn 2023, Vial et al., 2023).
Virion
Morphology
Orthohantavirions are pleomorphic in shape, mostly spherical of 80–160 nm in diameter (Huiskonen et al., 2010), and surrounded by a membrane envelope that is decorated with glycoprotein (GP) spikes composed of GN and GC subunits (Figure 1 Orthohantavirus). The virion envelope is 5 nm thick and is decorated with square shaped spikes, consisting of tetrameric GN-GC heterodimers, which protrude 10 nM from the membrane forming a surface lattice of local symmetry interrupted by empty membrane spots (Hung et al., 1983, Huiskonen et al., 2010, Battisti et al., 2011, Parvate et al., 2019, Serris et al., 2020, Guardado-Calvo and Rey 2021). Isolated ribonucleoprotein (RNP) complexes are composed of individual segment genomic RNA encapsidated by nucleoprotein (N) (Tao et al., 1987, Huiskonen et al., 2010, Battisti et al., 2011, Hepojoki et al., 2012, Guo et al., 2016, Arragain et al., 2019). Virions assemble and bud from the Golgi complex or cell surface membranes, depending on the virus (Goldsmith et al., 1995, Parvate et al., 2019).
 |
| Figure 1 Orthohantavirus. A, B) Electron micrographs of Tula virus particles adapted from (Huiskonen et al., 2010). C) Schematic illustration of an orthohantavirus particle. Shown is a spherical and enveloped (grey) particle with glycoprotein spikes (GN yellow, GC blue) inserted in a bilaminar lipid envelope. The S (small), M (medium), and L (large) RNP (ribonucleoprotein) complexes inside the particle consist of N (nucleoprotein, green) and L (large protein, pink). ViralZone image reproduced courtesy of SwissBioPics. |
Physicochemical and physical properties
Virion buoyant densities in sucrose and CsCl are 1.15–1.18 (White et al., 1982) and 1.20–1.21 g cm−3, respectively (Schmaljohn et al., 1983). Virions are sensitive to chemical inactivation with, for instance, methanol, paraformaldehyde, acetone/methanol, N-ethylmaleimide or detergents, as well as physical methods such as heat, gamma irradiation or UV irradiation (Kraus et al., 2005, Strandin et al., 2011, Elveborg et al., 2022).
Nucleic acid
Orthohantavirus genomes are linear tri-segmented RNA molecules of negative polarity. The genomes are about 11.1–12.3 kb in length (small [S] segment: 1.2–2.1 kb; medium [M] segment: 3.4–3.8 kb; large [L] segment: 6.5–6.6 kb).
Proteins
Orthohantaviruses typically express three structural proteins (Table 1 Orthohantavirus). The most abundant structural protein in an orthohantavirion is N, which encapsidates the orthohantaviral genomic segments. The least abundant protein is L, which mediates viral genome replication and transcription. Two glycoprotein subunits, GN and GC, mediate virion entry. GN and GC and non-structural proteins are encoded as a precursor polyprotein.
Table 1 Orthohantavirus. Location and function of orthohantavirus structural proteins.
| Protein | Location, mass, and function | Reference |
| Nucleoprotein (N) | Structural virion protein (about 48 kD). Oligomerizes and encapsidates orthohantaviral genomic segments and hence component of RNPs. Binds to L and GN | (Mir et al., 2010, Reuter and Krüger 2018, Arragain et al., 2019) |
| Nonstructural protein (NSs) | Non-structural protein encoded by some orhtohantaviruses. Has type I interferon inhibitory activity. May occur as multiple isoforms. | (Vera-Otarola et al., 2020, Binder et al., 2021) |
| Glycoprotein (GP) | Structural virion protein consisting of two subunits (GN: about 70-75 kD, GC: about 50-55 kD). Produced via proteolytic cleavage from the genome-encoded precursor GPC. Projects from virion membranes as tetrameric GP spikes composed of GN and GC heterodimers. GP mediates cell- receptor binding; as a class II fusion machine it induces virion-cell membrane fusion and, thereby cell entry. GN binds to N and RNA | (Spiropoulou et al., 2003, Tischler et al., 2005, Cifuentes-Muñoz et al., 2014, Li et al., 2016, Willensky et al., 2016, Rissanen et al., 2017, Zhu et al., 2018, Bignon et al., 2019, Sperber et al., 2019) |
| Large protein (L) | Structural virion protein (246 kD) with RdRP, helicase, and endoribonuclease domains. Component of the RNP inside virions. Binds to N and RNA. Oligomerizes and mediates transcription and replication of viral RNA segments. Mediates cap-snatching for viral mRNA capping. | (Kukkonen et al., 2005, Cheng et al., 2014, Rothenberger et al., 2016, Jeeva et al., 2019, Durieux Trouilleton et al., 2023) |
Genome organization and replication
The orthohantavirus genome consists of three negative-sense RNA molecules, termed S (small), M (medium), and L (large) (Figure 2 Orthohantavirus).
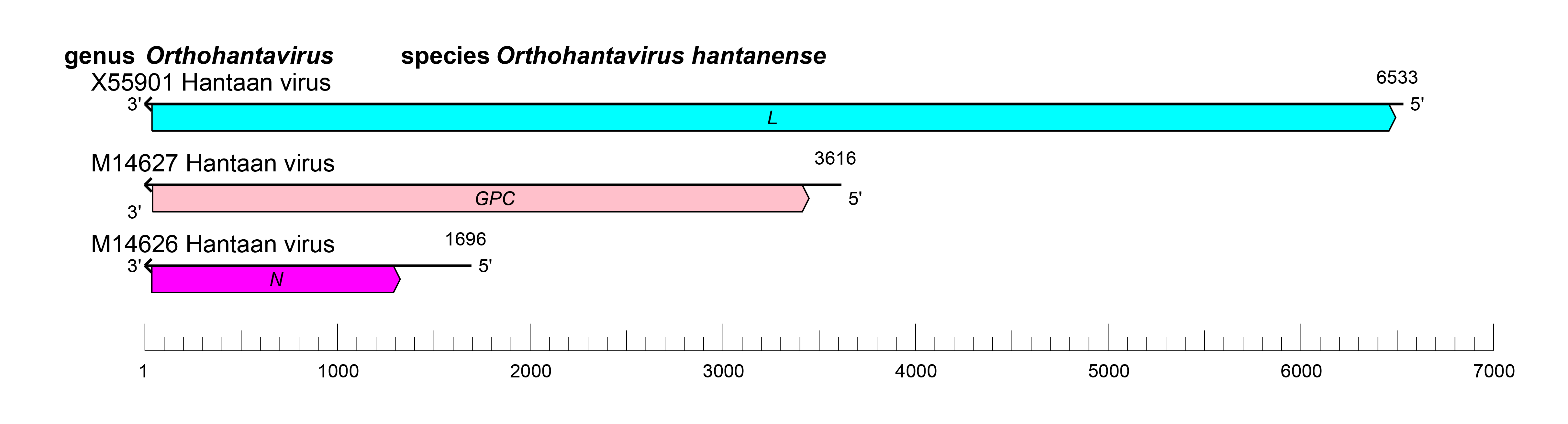 |
| Figure 2 Orthohantavirus. Schematic representation of orthohantavirus genome organization. |
Orthohantavirus genomic segments assume circular forms via non-covalent binding of complementary and highly conserved 3′- and 5′-terminal sequences. Genomic segment reassortment mostly occurs among viruses belonging to the same species but interspecies reassortment likely occurs occasionally (Klempa 2018, Petazzi et al., 2021).
Orthohantavirus infection starts with virion attachment, mediated by GN and GC, to cell-surface attachment factors and entry via the endosomal route (Shtanko et al., 2014) (Figure 3 Orthohantavirus).
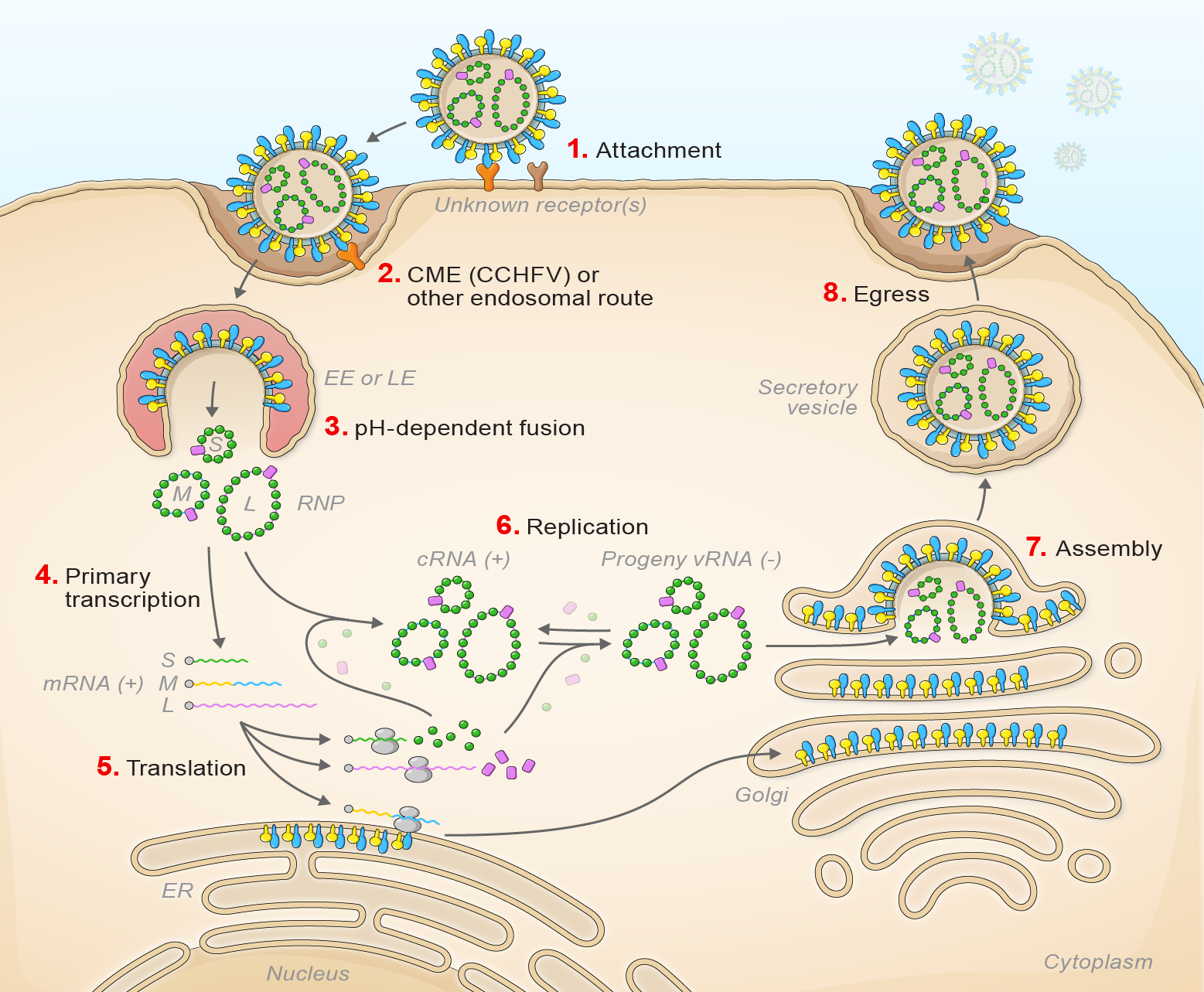 |
| Figure 3 Orthohantavirus. Lifecycle of orthohantaviruses. (1) Virion attachment; (2) virion uptake; (3) virion-cell membrane fusion; (4) transcription; (5) translation; (6) replication; (7) virion assembly; and (8) virion egress. |
After engaging a cellular receptor, protocadherin 1 in the case of American orthohantaviruses, virion-membrane fusion results in early or late endosomal release, depending on the virus, of the virion RNP complex into the cytoplasm in a pH-dependent manner (Jangra et al., 2018, Mittler et al., 2019, Torriani et al., 2019, Dieterle et al., 2021). During primary transcription L generates uncapped antigenomic RNA molecules that are then capped using host cell-derived capped primers (cap snatching). L and S segment-transcribed mRNAs are translated by free ribosomes (Garcin et al., 1995, Jeeva et al., 2019, Meier et al., 2021). M segment-transcribed mRNA is translated by membrane-bound ribosomes, with the expressed GPC which is likely co-translationally cleaved by cellular proteases to yield GN and GC. The synthesis of the antigenome by L protein serves as a template for genomic RNA replication. Secondary transcription amplifies the synthesis of mRNAs and genome replication. During morphogenesis, GN and GC accumulate in the Golgi apparatus, modified host membranes are acquired, and virions bud into the Golgi.
Biology
Genus Orthohantavirus includes 38 species for 60 distinct viruses. These viruses and their unclassified, potential relatives, subclinically, and often chronically, infect specific soricid and talpid eulipotyphla, and muroid and possibly dipodoid rodents (Jonsson et al., 2010, Wu et al., 2018, Chen et al., 2023, Kuhn and Schmaljohn 2023). Orthohantaviruses are overall distributed worldwide, but the individual geographic distribution is dependent on host range. At least some orthohantaviruses may not be as restricted to a particular host as originally thought, but may be carried by animals of multiple hosts (Goodfellow et al., 2021, Quizon et al., 2022, Gu et al., 2023).
Orthohantaviruses are the only hantavirids known to cause disease (all in humans) and all disease-causing viruses are rodent-borne. Human infection may occur after inhalation of aerosolized excreta or secreta of infected rodents or direct rodent contact. The two WHO-recognized orthohantavirus-caused diseases are hemorrhagic fever with renal syndrome (HFRS), caused by Asian and European viruses (in particular Dobrava virus [DOBV], Hantaan virus [HTNV], Puumala virus [PUUV] and Seoul virus [SEOV]), and hantavirus pulmonary syndrome (HPS), a frequently lethal disease caused by viruses from the Americas (in particular Andes virus [ANDV] and Sin Nombre virus [SNV]) (Vial et al., 2023).
The hamster model of wild-type ANDV infection is lethal and largely mimics human HPS (Hooper et al., 2001). After SNV infection in nonhuman primates, some animals develop an HPS-like disease, but cell passage of SNV can result in mutations that render the virus non-pathogenic in this model (Safronetz et al., 2014). Many rodent models result in a chronic, asymptomatic infection, and these have been used in early-stage therapeutic studies (Strandin et al., 2020). However, recent advances have been made in development of an HFRS mouse model using HTNV variants (Wei et al., 2022). Orthohantavirus animal models are reviewed in (Golden et al., 2015).
Antigenicity
There is considerable serum cross-reactivity between the nucleocapsids of hantavirids, but neutralizing antibodies targeting Gn/Gc are more specific. SNV was first identified as a hantavirid based on cross-reactivity of patient sera with other orthohantavirus antigens (1993). Similarly, rodents that carried SNV were identified in serosurveys using non-SNV orthohantavirus antigens, since SNV had not been isolated at the time (Childs et al., 1994).
Species demarcation criteria
Demarcation of species is based upon DivErsity pArtitioning by hieRarchical Clustering (DEmARC) analysis) using concatenated deduced S, M, and L segment expression product sequences (Laenen et al., 2019). Phylogenetic relationships across the genus have been estimated using maximum likelihood trees generated from complete N, GPC and L amino-acid sequences, and for N and GPC amino-acid sequences (Figure 2 Hantaviridae).
Related, unclassified viruses
Virus names and virus abbreviations are not official ICTV designations.
*Coding region sequence incomplete.

