Subfamily: Mammantavirinae
Genus: Loanvirus
Distinguishing features
Brno virus (BRNV) and Lóngquán virus (LQUV) are the only classified loanviruses. Both infect bats (Guo et al., 2013, Straková et al., 2017).
Virion
Virions are unknown.
Nucleic acid
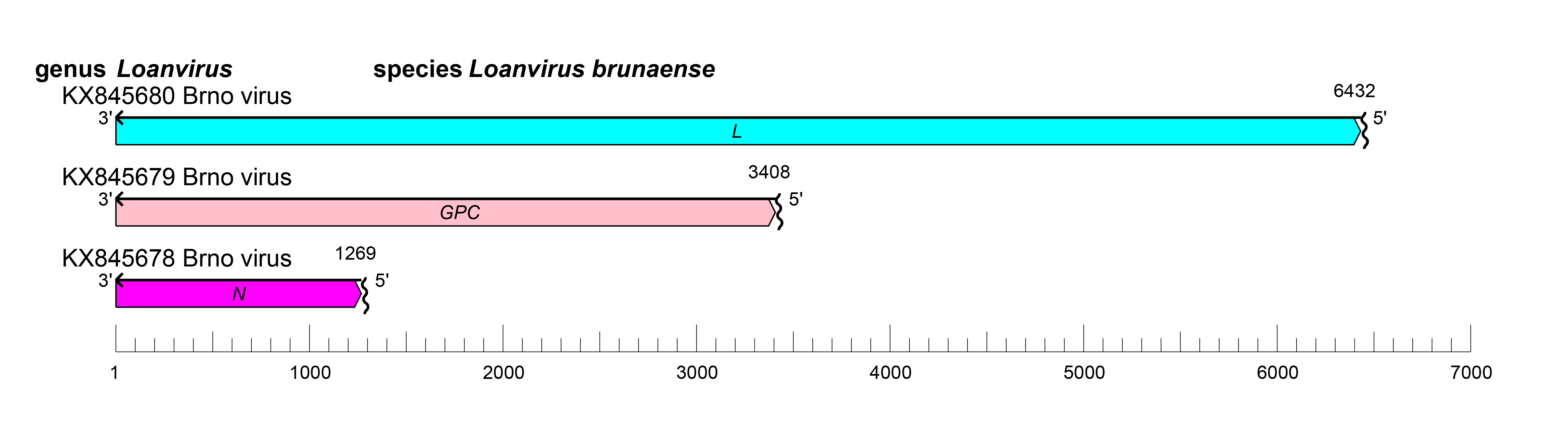
Loanviruses have tri-segmented negative-sense RNA genomes of about 11.9 kb (small [S] segment: 1.2 kb; medium [M] segment: 3.4–3.6 kb; large [L] segment: 6.2–6.4 kb) (Sumibcay et al., 2012, Weiss et al., 2012, Guo et al., 2013, Straková et al., 2017, Kouadio et al., 2020).
Proteins
Based on sequence data only, loanviruses likely express three structural proteins: nucleoprotein (N), glycoprotein precursor (GPC), and large protein (L) PMID: 28025098; PMID: 23408889.
Genome organization and replication
The S segment encodes N, the M segment encodes GPC, and the L segment encodes L (Figure 1 Loanvirus). Loanvirus genomic segments are expected to assume circular forms via non-covalent binding of complementary and conserved 3′ and 5′ terminal sequences.
 |
| Figure 1 Loanvirus. Schematic representation of loanvirus genome organization. The 5′ and 3′ ends of each segment (S, M and L) are, by analogy to other hantavirids, predicted to be complementary at their termini, likely promoting the formation of circular ribonucleoprotein complexes within the virion. |
Biology
BRNV infects noctules (vespertillionid Nyctalus noctule (Schreber, 1774)) in Austria, Czech Republic, Germany, and Poland (Straková et al., 2017, Dafalla et al., 2023). LQUV infects Chinese rufous horseshoe bats (rhinolophid Rhinolophus sinicus K. Andersen, 1905), Formosan lesser horseshoe bats (Rhinolophus monoceros Temminck, 1835), and intermediate horseshoe bats (Rhinolophus affinis Horsfield, 1823), in China (Guo et al., 2013). Unclassified, potential loanviruses infect vespertillionid bats in China and Côte d'Ivoire (Sumibcay et al., 2012, Guo et al., 2013), nycterid bats in Sierra Leone (Weiss et al., 2012), and murid rodents in Guinea (Kouadio et al., 2020). Replication-competent loanvirus isolates have not yet been obtained, and hence loanvirus biology remains to be elucidated.
Species demarcation criteria
Demarcation of species is based upon DivErsity pArtitioning by hieRarchical Clustering (DEmARC) analysis) using concatenated deduced S, M, and L segment expression product sequences (Laenen et al., 2019).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Huángpí virus | S: JX473273*; L: JX465369* | HUPV | (Guo et al., 2013) |
| Magboi virus | L: JN037851* | MGBV | (Weiss et al., 2012) |
| Méliandou hantavirus | L: MK726381* | MELV | (Kouadio et al., 2020) |
| Mouyassué virus | L: JQ287716* | MOUV | (Sumibcay et al., 2012) |
| Ponan loanvirus | S: MN597047; M: MN597045*; L: MN597043 |
Virus names and virus abbreviations are not official ICTV designations.
*Coding region sequence incomplete.

