Family: Hantaviridae
Steven B. Bradfute, Charles H. Calisher, Boris Klempa, Jonas Klingström, Jens H. Kuhn, Lies Laenen, Nicole D. Tischler and Piet Maes
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Piet Maes (piet.maes@kuleuven.be)
Edited by: Holly Hughes and Evelien Adriaenssens
Posted: March 2024, updated June 2024
Summary
Hantaviridae is a family for negative-sense RNA viruses with genomes of about 10.5–14.6 kb (Table 1 Hantaviridae). The family includes eight genera (Actinovirus and Percilovirus (subfamily Actantavirinae), Agnathovirus (subfamily Agantavirinae), Loanvirus, Mobatvirus, Orthohantavirus and Thottimvirus (subfamily Mammantavirinae), and Reptillovirus (subfamily Repantavirinae). These viruses are maintained in and/or transmitted by small mammals, but also by fish and reptiles. Several orthohantaviruses can infect humans, causing mild, severe, and sometimes-fatal diseases. Hantavirids produce enveloped virions containing three single-stranded RNA segments with open reading frames (ORFs) that encode a nucleoprotein (N), a glycoprotein precursor (GPC), and large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain.
Table 1 Hantaviridae. Characteristics of members of the family Hantaviridae.
| Characteristic | Description* |
| Example | Hantaan virus (S segment: M14626; M segment: M14627; L segment: X55901), species Orthohantavirus hantanense |
| Virion | Enveloped, pleomorphic virions 80–160 nm in diameter with heterodimeric surface spikes |
| Genome | Three single-stranded RNA molecules (segments): small (S; 1.0–3.0 kb), medium (M; 3.4–4.8 kb), and large (L; 5.3–6.8 kb) |
| Replication | Ribonucleoprotein (RNP) complexes containing anti-genomic RNA serve as coding templates for synthesis of genomic RNA |
| Translation | Proteins are produced from capped and non-polyadenylated mRNAs. The 5′-cap structure is obtained via cap-snatching from cellular mRNAs |
| Host range | Fish (actinoviruses, agnathoviruses, perciloviruses), mammals (orthohantaviruses), and reptiles (reptilloviruses) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Elliovirales; the family includes four subfamilies, eight genera, and 55 species |
* mostly based on experiments with mammalian orthohantaviruses
Viruses assigned to each of the seven genera form genus-specifc monophyletic clades based on phylogenetic analysis of concatenated L protein/RdRP, glycoprotein and N sequences. Viruses from all seven genera share one or more of the following characteristics: (i) enveloped pleomorphic virions; (ii) tri-segmented, negative-sense RNA genome without polyadenylated tracts at the 3′-end; (iii) genomic sequence complementarity at the 5′- and 3′-ends; and (iv) capped but not polyadenylated virus mRNAs (Maes et al., 2004).
Piscine Hosts
Genus Actinovirus. Viruses in this genus infect actinopterygian (percid, ogcocephalid, and triacanthodid, and possibly goblid and melanotaeniid) fish.
Genus Agnathovirus. Viruses in this genus infect agnathan (myxinid) fish.
Genus Percilovirus. Viruses in this genus infect actinopterygian (gobiid, lophiid) fish.
Mammalian Hosts
Genus Loanvirus. Viruses in this genus infect rhinolophid, verspertillionid, and possibly nycterid bats and possibly murid rodents.
Genus Mobatvirus. Viruses in this genus infect emballonurid, hipposiderid, pteropodid and possibly molossid, and vespertillionid bats and soricid and talpid eulipotyphla.
Genus Orthohantavirus. Viruses assigned to this genus infect soricid and talpid eulipotyphla and muroid and possibly dipodoid rodents.
Genus Thottimvirus. Viruses in this genus infect soricid and possibly talpid eulipotyphla.
Reptilian Hosts
Genus Reptillovirus. Viruses in this genus infect gekkonid and possibly scincid reptiles.
Virion
Morphology
Only known for members of the genus Orthohantavirus (see Orthohantavirus genus page).
Physicochemical and physical properties
Only known for members of the genus Orthohantavirus (see Orthohantavirus genus page).
Nucleic acid
Hantavirids have tri-segmented, negative-sense RNA genomes (S, M, and L segments). The viral mRNAs are not polyadenylated and contain a 5′-methylated cap and several non-templated nucleotides at the 5′-end that are derived from host cell mRNAs via cap snatching (Schmaljohn et al., 1985, Jonsson and Schmaljohn 2001, Vaheri et al., 2013). Further information is only known for members of the genus Orthohantavirus (see Orthohantavirus genus page).
Proteins
Hantavirids typically express three structural proteins. The most abundant structural protein in a hantavirion is N, which encapsidates the hantaviral genomic segments. The least abundant protein is L, which mediates viral genome replication and transcription. Two glycoprotein subunits, GN and GC, are produced from GPC and mediate entry of hantavirions (Schmaljohn et al., 1985, Spiropoulou 2001, Vaheri et al., 2013). Specifics are only known for members of the genus Orthohantavirus (see Orthohantavirus genus page).
Lipids
Not reported but are likely derived from the Golgi apparatus and/or the host cell plasma membrane and therefore likely composed of phospholipids, glycolipids, fatty acids, and sterols.
Carbohydrates
Only known for members of the genus Orthohantavirus (see Orthohantavirus genus page).
Genome organization and replication
S RNA encodes N and in some viruses NSs (Jääskeläinen et al., 2007, Vera-Otarola et al., 2012), M RNA encodes GPC, and the L RNA encodes L with its RdRP, helicase, and endonuclease domains (Jonsson and Schmaljohn 2001) (Figure 1 Hantaviridae). Specifics are only known for members of the genus Orthohantavirus (see Orthohantavirus genus page).
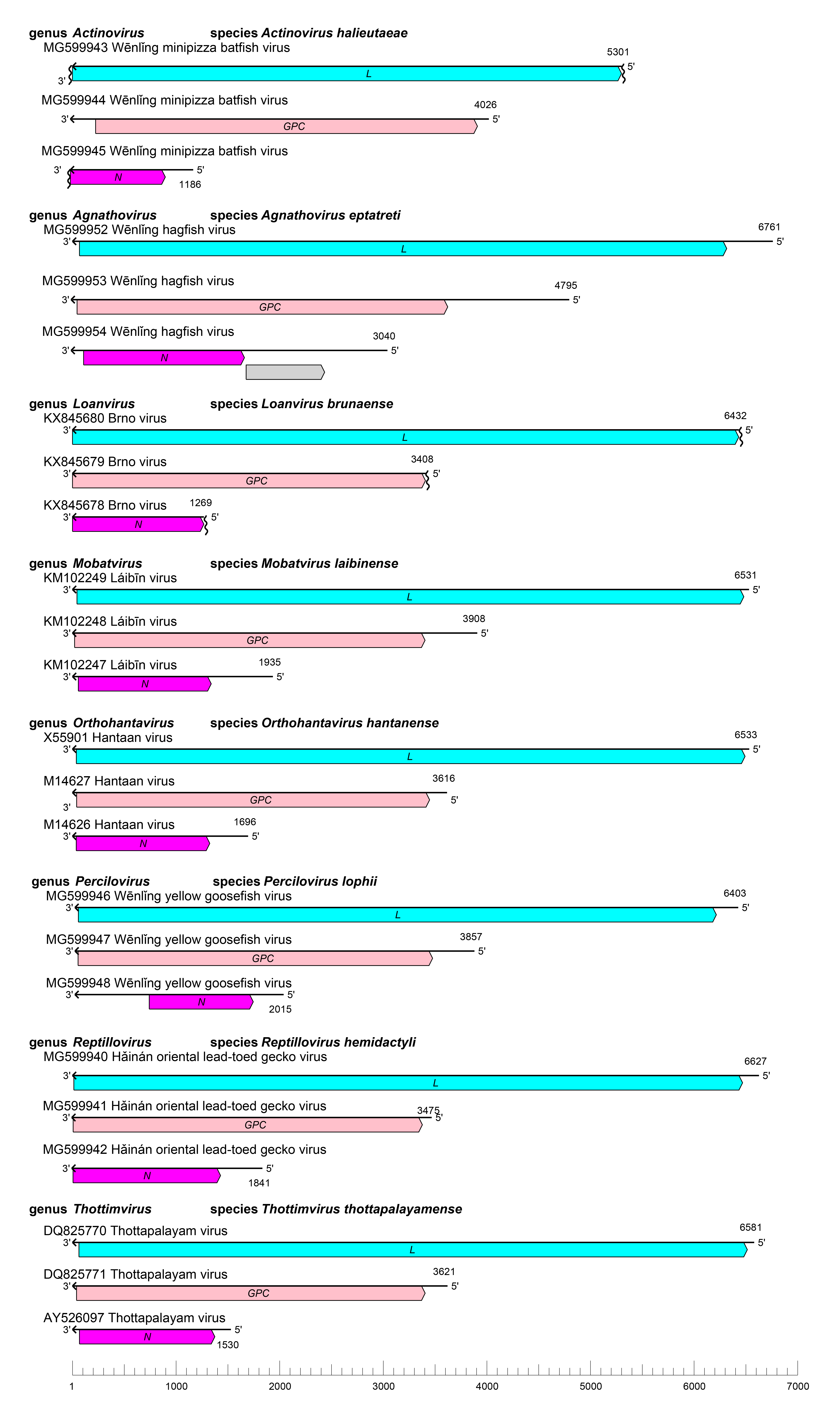 |
| Figure 1 Hantaviridae. Genome organization of representative hantaviruses. The 5′- and 3′- ends of all segments are complementary at their termini, likely promoting the formation of panhandle RNP complexes within the virion. GPC, glycoprotein precursor gene; L, large (protein) gene; N, nucleoprotein gene. |
Biology
Hantavirids have been found in actinopterygian (gobiid, lophiid, percid, ogcocephalid, and triacanthodid, and possibly goblid and melanotaeniid) and agnathan (myxinid) fish; in emballonurid, hipposiderid, pteropodid, rhinolophid, verspertillionid, and possibly molossid and nycterid bats; in soricid and talpid eulipotyphla; in muroid and possibly dipodoid rodents; in gekkonid and possibly scincid reptiles; and potentially in coleopteran and plectopteran insects (see genus pages for details). Only orthohantaviruses are known to cause diseases (see Orthohantavirus genus page).
Antigenicity
Systematic antigenicity studies have only been reported for orthohantaviruses (see Orthohantavirus genus page).
Derivation of names
Actantavirinae: after the fish class Actinopterygii and hantavirus
Actinovirus: after the fish class Actinopterygii
Agantavirinae: after the fish infraphylum Agnatha and hantavirus
Agnathovirus: after the fish infraphylum Agnatha
andesense: after the Andes mountain range, South America
artybashense: after Artybash (Артыбаш), Altai Republic (Республика Алтай), Russia
asamaense: after the Asama River, Japan
asikkalaense: after Asikkala, Finland
bayoui: after the Latin bayouus, meaning bayou
bernense: after Bern, Switzerland
boweense: after Bowé, Guinea
brugesense: after Bruges, Belgium
brunaense: after the Latin Bruna, meaning Brno, Czech Republic
caobangense: after Cao Bằng Province, Vietnam
carrizalense: after Carrizal virus
chocloense: after the cantina (bar) “El Choclo”, Panama
dabieshanense: after 大別山 (Dàbiéshān), China
dakrongense: after Đakrông, Quảng Trị Province, Vietnam
delgaditoense: after Caño Delgadito, Venezuela
dobravaense: after Dobrava in Slovenia
eptatreti: after the fish genus Eptatretus
fugongense: after 福贡县 (Fúgòng County), China
gobbii: after wolfsnout goby (family Gobiidae)
halieutaeae: after the fish genus Halieutaea
hantanense: after 한탄강 (Hantan River), South Korea
Hantaviridae: after Hantaan virus
hemidactyli: after the reptilian genus Hemidactylus
imjinense: after 임진강 (Imjin river), South Korea
jejuense: after 제주도 (Jeju Island), South Korea
kenkemeense: after Кенкеме (Kenkeme river), Russia
khabarovskense: after Хабаровск (Khabarovsk), Russia
laibinense: after 来宾 (Láibīn), China
lankaense: after Sri Lanka
lenaense: after Ле́на (Lena river), Russia
Loanvirus: after 龙泉 (Lóngquán), China
longquanense: after 龙泉 (Lóngquán), China
lophii: after the fish genus Lophius
luxiense: after 泸溪县 (Lúxī County), China
Mammantavirinae: after the animal class Mammalia and hantavirus
mamorense: after Rio Mamoré virus
maporalense: after Hato Maporal farm, Venezuela
Mobatvirus: after mole and bat
montanoense: after a geographical feature or location (Montaño) in or near Leonardo Bravo municipality, Mexico
nigrorivense: after the Latin nigro, meaning black and the Latin rivus, meaning river
novaense: after Nova, Hungary
Orthohantavirus: from the Ancient Greek ὀρθός (orthós), meaning straight, right, proper and Hantaan virus
Percilovirus: from the host order names Perciformes and Lophiiformes
prospectense: after Prospect Hill, USA
puumalaense: after Puumala Municipality, Finland
quezonense: after Quezon Memorial National Park, Philippines
Repantavirinae: after the animal class Reptilia and hantavirus
Reptillovirus: after the animal class Reptilia
robinaense: after Robina, Australia
rockportense: after Rockport, USA
sangassouense: after Sangassou, Guinea
seoulense: after Seoul, South Korea
sinnombreense: after Sin Nombre canyon, USA
tatenalense: after historic Tatenale, England
thailandense: after Thailand
thottapalayamense: after Thottapalayam, India
Thottimvirus: after Thottopalayam virus
tigrayense: after ትግራይ (Tigray Region), Ethiopia
triacanthodis: after the fish genus Triacanthodes
tulaense: after Тула (Tula), Russia
xuansonense: after Xuân Sơn, Vietnam
wufangense: after after Wùfēng (吾锋), Fújiàn Province (福建省), China
Subfamily, genus and species demarcation criteria
The availability of at least coding-complete sequences of all three genome segments may be sufficient for hantavirid classification in the absence of a cultured isolate. Demarcation of genera is based upon DivErsity pArtitioning by hieRarchical Clustering (DEmARC) analysis) using concatenated deduced S, M, and L segment expression product sequences (Hantaviridae: current classification and future perspectives, Viruses, 11, 9, e0788">Laenen et al., 2019). DEmARC analysis gave a frequency distribution of Pairwise Evolutionary Distance (PED) values of which the threshold of 0.1 gave an optimal clustering cost of zero and is used as the hantavirid species demarcation criterium. Genera are demarked by a PED-value threshold of 0.95 and subfamilies are demarcated based on their distinct clustering in a Bayesian maximum clade credibility tree and a PED-value threshold of 3.5.
Relationships within the family
Phylogenetic relationships across the family have been estimated using Bayesian maximum clade credibility trees generated from complete N, GPC, and L amino-acid sequences; similar topologies are obtained with concatenated N and GPC sequences (Figure 2 Hantaviridae).
 |
| Figure 2 Hantaviridae. Maximum likelihood phylogenetic tree inferred using IQ-TREE. The tree was constructed based on 53 concatenated S and M segment (amino acid) sequences using the LG+R9 model of sequence evolution. Branch lengths represent the estimated number of substitutions per site. Bootstrap support values (>70%) are indicated at nodes, reflecting the degree of confidence in the branching pattern. |
Relationships with other taxa
Hantavirids are closely related to bunyaviral crulivirids, fimovirids, peribunyavirids, phasmavirids, tulasvirids, and tospovirids (Wolf et al., 2018, Herath et al., 2020).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| coleopteran hanta-related virus OKIAV221 | L: MT153497 | (Käfer et al., 2019) | |
| plecopteran hanta-related virus OKIAV215 | L: MT153434 | (Käfer et al., 2019) |
Virus names and virus abbreviations are not official ICTV designations.
Additional unclassified hantavirids that are probable members of established genera are listed under individual genus descriptions.

