Family: Filoviridae
Genus: Thamnovirus
Distinguishing features
This genus includes three classified viruses: Fiwi virus (FIWIV), Kander virus (KNDV), and Huángjiāo virus (HUJV). Like oblaviruses and striaviruses, but unlike cuevaviruses, dianloviruses, orthoebolaviruses, orthomarburgviruses, and tapjoviruses, thamnoviruses infect fish. Notably, thamnovirus genomes encode at least two proteins without obvious homologs in other filovirid genera and do not encode a matrix protein (VP40) or a ribonucleoprotein (RNP) complex-associated protein (VP24) (Shi et al., 2018, Hume and Mühlberger 2019, Hierweger et al., 2021).
Virion
Virions are unknown.
Nucleic acid
Virions are assumed to contain one or several copies of the linear negative-sense RNA genome that are encapsidated independently.
Proteins
Thamnoviruses likely express at least seven proteins, of which at least five (nucleoprotein [NP], polymerase cofactor [VP35], glycoprotein [GP1,2], transcriptional activator [VP30], and large protein [L]) are homologs of proteins expressed by other filovirids (Shi et al., 2018, Hume and Mühlberger 2019, Hierweger et al., 2021). After VP40, the second most abundant structural protein in a thamnovirion is assumed to be NP, which encapsidates the striavirus genome. The least abundant protein is assumed to be L, which mediates thamnovirus genome replication and transcription.
Genome organization and replication
The thamnovirus genome has the gene order 3′-U1-NP-VP35-U2-GP-VP30-L-5′ (Shi et al., 2018, Hume and Mühlberger 2019, Hierweger et al., 2021), with U denoting genes expressing proteins of unknown function (Figure 1.Thamnovirus). The undetermined extragenic sequences at the extreme 3′-end (leader) and 5′-end (trailer) of the genome are assumed to be conserved and partially complementary. The transcriptional initiation sites are unclear but relatively conserved likely termination (polyadenylation) sites have been identified (Hume and Mühlberger 2019).
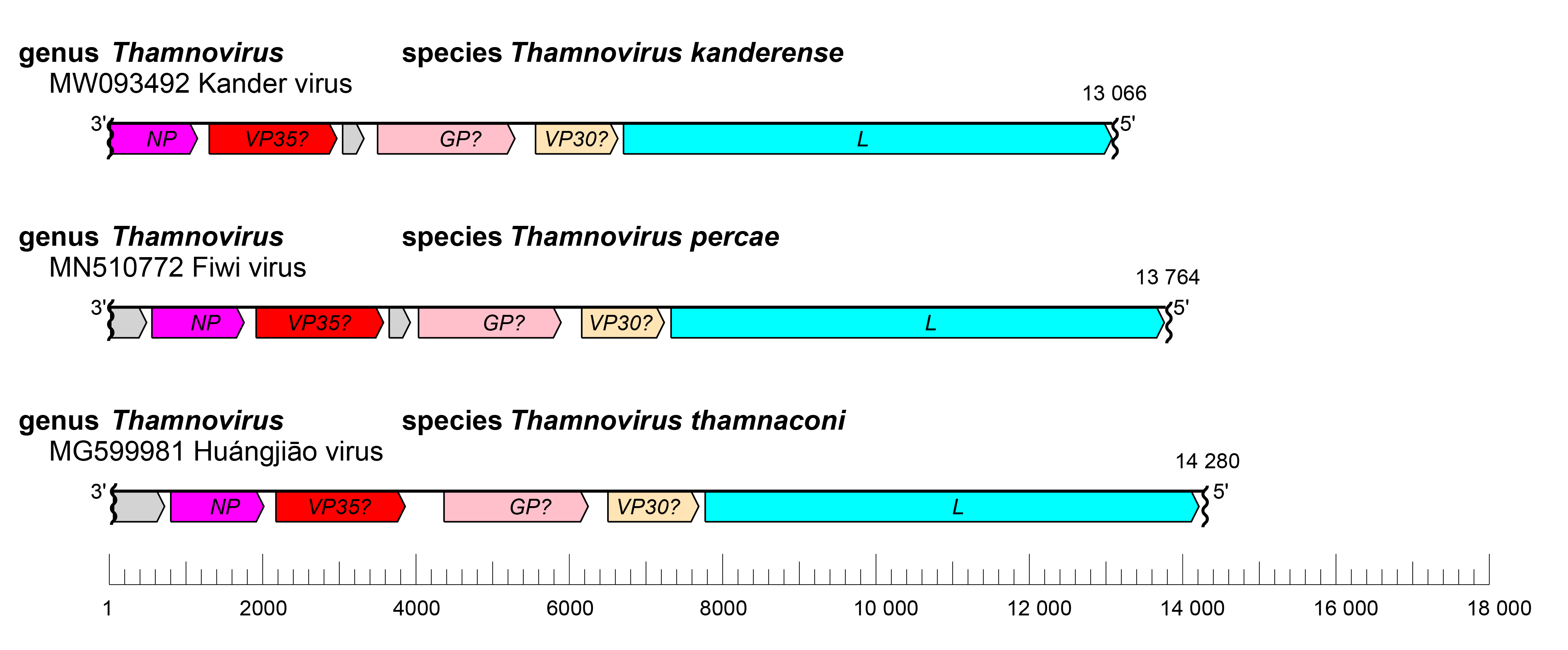 |
| Figure 1.Thamnovirus. Schematic representation of thamnovirus genome organization, drawn to scale. GP, glycoprotein gene; L, large protein gene; NP, nucleoprotein gene; U1/2, genes encoding proteins of unknown function; VP30, transcriptional activator gene; VP35, polymerase co-factor gene. Wavy lines indicate incomplete genome ends. |
The replication strategy of thamnoviruses remains to be studied.
Biology
Thamnoviruses were discovered by high-throughput sequencing of samples taken from greenfin horse-faced filefish (monacanthid Thamnaconus septentrionalis (Günther, 1874)), captured by fishing trawlers in the East China Sea (Shi et al., 2018), and by high-throughput sequencing of samples taken from farmed diseased European perch (percid Perca fluviatilis Linnaeus, 1758) imported to Switzerland from Germany (Hume and Mühlberger 2019, Hierweger et al., 2021).
Species demarcation criteria
The genus currently includes three species. PAirwise Sequence Comparison (PASC) using coding-complete thamnovirus genomes is the primary tool for thamnovirus species demarcation. Genomic sequences of thamnoviruses of different species differ from each other by ≥23% (Bào et al., 2017).

