Family: Filoviridae
Genus: Cuevavirus
Distinguishing features
Lloviu virus (LLOV) is the only currently classified cuevavirus. Like dianloviruses, orthomarburgviruses, Bombali virus (BOMV) and possibly other orthoebolaviruses cuevaviruses infect bats (Negredo et al., 2011, Kemenesi et al., 2018, Kemenesi et al., 2022, Tóth et al., 2023).
Virion
Morphology
Similar to particles produced by orthoebolaviruses and orthomarburgviruses, cuevavirions are filamentous in shape, but can also be branched or 6 shaped (Hume et al., 2022) (Figure 1.Cuevavirus). Virions are composed of a central core formed by a helical ribonucleoprotein (RNP) complex, surrounded by a matrix protein (VP40) layer and a lipid envelope derived from the host cell plasma membrane that contains glycoprotein (GP1.2) complexes (Maruyama et al., 2014, Hume et al., 2022, Hu et al., 2023).
 |
| Figure 1.Cuevavirus. Electron microscopy (EM) of LLOV-infected cells and virions. (Left) Transmission EM of LLOV particles (top panel, arrow) and cross section of LLOV inclusions (bottom panel). Asterisks indicate nucleocapsids within the LLOV inclusions. (right) EM of negatively-stained virions. Figure reproduced from (Hume et al., 2022) under Creative Commons Attribution (CC BY) license. |
Nucleic acid
Cuevavirus genomes are linear non-segmented RNA molecules of negative polarity. Genomic RNA is about 19 kb (Negredo et al., 2011, Hume et al., 2022, Tóth et al., 2023). and likely uncapped and not polyadenylated.
Proteins
Cuevaviruses express seven structural proteins (Table 1.Cuevavirus), all of which are homologous to those of other mammalian filovirids (dianloviruses, orthoebolaviruses, and orthomarburgviruses) (Kämper et al., 2019). After VP40, the second most abundant structural protein in virions is assumed to be the nucleoprotein (NP), which encapsidates the cuevavirus genome. The least abundant protein is assumed to be the large protein (L), which mediates cuevavirus genome replication and transcription via its RNA-directed RNA polymerase (RdRP) domain. The cuevavirus RNP complex likely consists of NP, VP24, polymerase cofactor (VP35), transcriptional activator (VP30), and L. These RNP complexes associate with the matrix protein (VP40), which lines the inner side of the virion membrane and GP1 and GP2, which form globular spikes on the outside of the virion membrane. Similar to orthoebolaviruses, but unlike dianloviruses and orthomarburgviruses, cuevaviruses express GP1 and GP2 via co-transcriptional editing and also express three soluble glycoproteins from the GP gene (sGP, ssGP, and Δ-peptide) (Negredo et al., 2011, Manhart et al., 2018).
Table 1.Cuevavirus. Location and functions of cuevavirus structural proteins.
| Protein | Encoding gene | Characteristics | Function | Reference |
|---|---|---|---|---|
| Nucleoprotein (NP) | 1 (NP) | RNP complex component; likely consists of two distinct functional modules; homo oligomerizes to form helical polymers; binds to genomic and antigenomic RNA, VP35, VP40, VP30, and VP24 | Nucleocapsid and cellular inclusion body formation; encapsidation of cuevavirus genome and antigenome; genome replication and transcription | (Manhart et al., 2018, Kämper et al., 2019) |
| Polymerase cofactor (VP35) | 2 (VP35) | RNP complex component; homo oligomer; binds to double stranded RNA, NP, and L | Replicase transcriptase cofactor; inhibits interferon regulatory factor 3 phosphorylation, IFNA1/B1 production, and protein kinase R phosphorylation | (Negredo et al., 2011, Feagins and Basler 2015, Manhart et al., 2018, Kämper et al., 2019) |
| Matrix protein (VP40) | 3 (VP40) | Likely consists of two distinct functional modules; homo oligomerizes to form dimers and circular hexamers and octamers; binds single-stranded RNA, VP35; hydrophobic; membrane associated; contains three late budding motifs; binds to NEDD4 and TSG101; binds to tubulin alpha and is ubiquitinylated | Matrix component; regulation of genome transcription and replication; regulation of virion morphogenesis and egress | (Negredo et al., 2011, Kämper et al., 2019) |
| Secreted glycoprotein (sGP) | 4 (GP) | Likely secreted as a parallel homo dimer; likely N glycosylated, C mannosylated, sialylated | Unknown | |
| Glycoprotein ( GP1 and GP2) | 4 (GP) | Type I transmembrane and class I fusion protein; cleaved to GP1 and GP2 subunits that heterodimerize; mature protein is a trimer of GP1 and GP2 heterodimers; inserts into membranes; heavily N- and O glycosylated | Virion adsorption to cuevavirus-susceptible cells via cellular attachment factors; determines cuevavirus cell and tissue tropism; induction of virus-cell membrane fusion subsequent to endolysosomal binding to NPC1; inhibits innate immune response by interfering with BST2 | (Negredo et al., 2011, Maruyama et al., 2014, Ng et al., 2014, Brinkmann et al., 2016, Kämper et al., 2019) |
| Secondary secreted glycoprotein (ssGP) | 4 (GP) | Nonstructural; secreted as a glycosylated monomer | Unknown | |
| Δ-peptide | 4 (GP) | Nonstructural; secreted; largely unstructured; likely O glycosylated and sialylated | Hypothesized to suppress filovirid superinfection | (Radoshitzky et al., 2011) |
| Transcriptional activator (VP30) | 5 (VP30) | RNP complex component; hexameric zinc finger protein; binds single stranded RNA, NP, and L | Transcription initiation at the NP gene start; potentially reinitiation and antitermination | (Manhart et al., 2018, Kämper et al., 2019, Sun et al., 2022) |
| RNP complex associated protein (VP24) | 6 (VP24) | Likely RNP complex component; homo tetramerizes; hydrophobic and membrane-associated | Regulation of genome transcription and replication; regulation of virion morphogenesis and egress; inhibits tyrosine phosphorylated STAT1 binding to KPNA5, STAT1 nuclear accumulation, both RIGI- and IFIH1-stimulated IFNB and IFNL1 promoter activation, and IFNA-induced MX1 and IFITM3 promoter activation | (Negredo et al., 2011, Feagins and Basler 2015, Kämper et al., 2019, He et al., 2021, Khan et al., 2023) |
| Large protein (L) | 6 (L) | RNP complex component; homo dimerizes; binds to genomic and antigenomic RNA, VP35, and VP30; mRNA capping enzyme | Genome replication and mRNA transcription; co transcriptional editing | (Manhart et al., 2018) |
BST2, bone marrow stromal antigen 2; IFIH1, interferon induced with helicase C domain 1 (formerly MDA5); IFNA, interferon alpha; IFNB, interferon beta; IFNL, interferon lambda; KPNA5, karyopherin subunit alpha 5; MX1, MX dynamin like GTPAse 1 (formerly MxA); NEDD4, NEDD4 E3 ubiquitin protein ligase; NPC1, NPC intracellular cholesterol transporter 1; RNP, ribonucleoprotein; STAT1, signal transducer and activator of transcription 1; TSG101, tumor susceptibility 101; VP, virus protein
Genome organization and replication
The cuevavirus genome has the gene order 3′-NP-VP35-VP40-GP-VP30-VP24-L-5′ (Figure 2.Cuevavirus). The undetermined extragenic sequences at the extreme 3′-end (leader) and 5′-end (trailer) of the genome are assumed to be conserved and to be partially complementary. Genes are flanked by conserved transcriptional initiation and termination (polyadenylation) sites. The cuevavirus transcriptional initiation site sequence is identical to that of orthoebolaviruses, but the transcriptional termination sequence is unique (3′-CUUCUU(A/G)UAAUU-5′); the VP24 transcriptional termination sequence and the L transcriptional termination sequence have not yet been identified.. Most cuevavirus genes overlap. Most of these overlaps are extremely short and limited to the highly conserved pentamer. Most genes possess relatively long 3′-end and 5′-end noncoding regions. Similar to orthoebolaviruses, but contrary to dianloviruses and orthomarburgviruses, the GP genes of cuevaviruses possess three overlapping open reading frames (ORFs) that can be joined through co transcriptional polymerase stuttering (Negredo et al., 2011, Tóth et al., 2023).
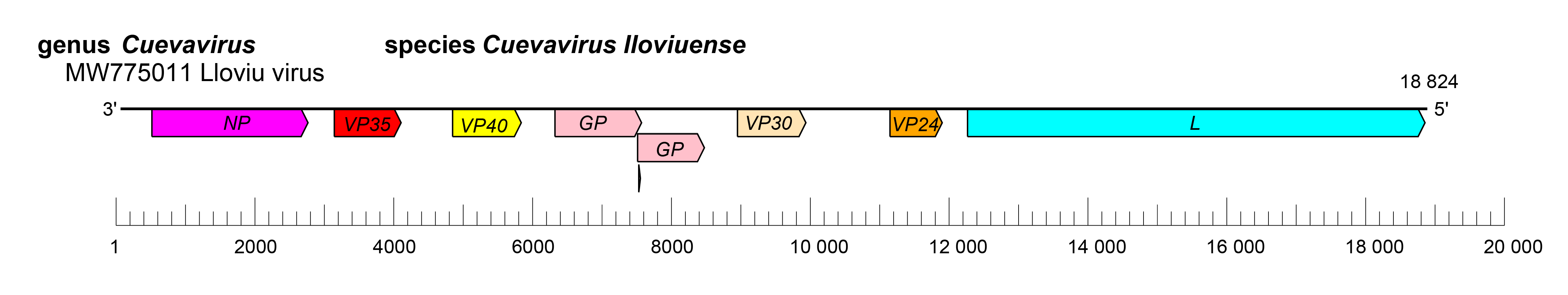 |
| Figure 2.Cuevavirus. Schematic representation of cuevavirus genome organization. Genome is drawn to scale. |
The replication strategy of cuevaviruses is highly similar to that of orthoebolaviruses and reminiscent of that of dianloviruses and orthomarburgviruses (Manhart et al., 2018, Kämper et al., 2019, Hume et al., 2022). Cuevavirions are assumed to associate with attachment factors at the plasma membrane that mediate infection by endocytosis. Cuevavirus GP1 and GP2 mediate cell surface C-type lectin binding and subsequent low-pH-dependent fusion into endosomes. Cathepsin L cleavage is required for GP1 and GP2 binding to the endosomal receptor NPC intracellular cholesterol transporter 1 (NPC1) (Maruyama et al., 2014, Ng et al., 2014, Takadate et al., 2020), which is also used by dianloviruses, orthoebolaviruses, and orthomarburgviruses. Uncoating is presumed to occur in a manner analogous to that of other mononegaviruses. Cuevavirus transcription and genome replication likely take place in the cytoplasm and, in general, follow the models for members of the families Paramyxoviridae and Pneumoviridae. Transcription starts at the conserved transcriptional initiation site, and polyadenylation occurs at a stretch of uridine residues within the transcriptional termination site. The 5′-terminal non-coding sequences favor hairpin like structures for all mRNAs. Replication involves the synthesis of full-length positive-sense copies (antigenomes) (Manhart et al., 2018, Hume et al., 2022). During infection, it is assumed that massive amounts of nucleocapsids accumulate intracellularly and form intracytoplasmic inclusion bodies. Virions are likely released via budding from plasma membranes (Figure 3.Filoviridae).
Biology
Cuevaviruses were discovered in 2002 by high-throughput sequencing of samples taken from dead Schreibers’s long-fingered bats (Miniopterus schreibersii Kuhl, 1817) in Spain (Negredo et al., 2011). They were re-discovered in 2016–2023 in live and dead Schreibers’s long-fingered bats collected in Hungary and Italy (Kemenesi et al., 2018, Sun et al., 2022, Tóth et al., 2023).
Antigenicity
Initial studies indicate that LLOV is antigenically distinct from other filovirids (Maruyama et al., 2014, Ramírez de Arellano et al., 2019).
Species demarcation criteria
The genus currently includes only a single species.

