Family: Filoviridae
Genus: Dianlovirus
Distinguishing features
Měnglà virus (MLAV) is the only currently classified dianlovirus. Like cuevaviruses, orthomarburgviruses, and Bombali virus (BOMV) and possibly other orthoebolaviruses, dianloviruses infect bats. Dianlovirus genomes are highly reminiscent in organization of orthomarburgvirus genomes but contain four rather than only one gene overlap (Yang et al., 2019, Makenov et al., 2023).
Nucleic acid
The dianlovirus genomes is a linear non-segmented RNA molecule of negative polarity. Genomic RNA is about 18.5 kb (Yang et al., 2019) and is likely uncapped and not polyadenylated.
Proteins
Dianloviruses express seven structural proteins, all of which are homologous to those of cuevaviruses, orthoebolaviruses, and orthomarburgviruses (Table 1.Dianlovirus). After VP40, the second most abundant structural protein in virions is assumed to be the nucleoprotein (NP), which encapsidates the dianlovirus genome. The least abundant protein is assumed to be the large protein (L), which mediates dianlovirus genome replication and transcription via its RNA-directed RNA polymerase (RdRP) domain. The dianlovirus ribonucleoprotein (RNP) complex likely consists of NP, RNP complex-associated protein (VP24), polymerase cofactor (VP35), transcriptional activator (VP30), and L. These RNP complexes associate with the matrix protein (VP40), which lines the inner side of the virion membrane and GP1 and GP2, which form globular spikes on the outside of the virion membrane (Yang et al., 2019).
Table 1.Dianlovirus. Location and functions of dianlovirus structural proteins.
| Protein | Encoding gene | Characteristics | Function | Reference |
|---|---|---|---|---|
| Nucleoprotein (NP) | 1 (NP) | RNP complex component; likely consists of two distinct functional modules; homo-oligomerizes to form helical polymers; binds to genomic and antigenomic RNA, VP35, VP40, VP30, and VP24 | Nucleocapsid and cellular inclusion body formation; encapsidation of dianlovirus genome and antigenome; genome replication and transcription | (Yang et al., 2019) |
| Polymerase cofactor (VP35) | 2 (VP35) | RNP complex component; homo oligomer; binds to double stranded RNA, NP, and L | Replicase transcriptase cofactor; inhibits virus-induced activation of the IFNB promoter and IRF3 phosphorylation, inhibits IFNA1/B1 production, and protein kinase R and PRKRA phosphorylation | (Yang et al., 2019, Williams et al., 2020) |
| Matrix protein (VP40) | 3 (VP40) | Likely consists of two distinct functional modules; homo-oligomerizes to form dimers and circular hexamers and octamers; binds single-stranded RNA, VP35; hydrophobic; membrane associated; contains one late budding motif; binds NEDD4 and TSG101 | Matrix component; regulation of genome transcription and replication; regulation of virion morphogenesis and egress; inhibits JAK STAT pathway | (Yang et al., 2019, Williams et al., 2020) |
| Glycoprotein (GP1,2) | 4 (GP) | Type I transmembrane and class I fusion protein; cleaved to GP1 and GP2 subunits that heterodimerize; mature protein is a trimer of GP1 and GP2 heterodimers; inserts into membranes; heavily N- and O glycosylated | Virion adsorption to dianlovirus susceptible cells via cellular attachment factors; determines dianlovirus cell and tissue tropism; induction of virus cell membrane fusion subsequent to endolysosomal binding to NPC1 | (Yang et al., 2019) |
| Transcriptional activator (VP30) | 5 (VP30) | RNP complex component; hexameric zinc finger protein; binds single stranded RNA, NP, and L | Potentially transcription initiation, reinitiation, and/or antitermination | (Yang et al., 2019, Dong et al., 2020) |
| RNP complex-associated protein (VP24) | 6 (VP24) | Likely RNP complex component; homo tetramerizes; hydrophobic and membrane associated | Regulation of genome transcription and replication; regulation of virion morphogenesis and egress; inhibits IFN induced MX1 and IFITM3 promoter activation | (Yang et al., 2019, Williams et al., 2020, Khan et al., 2023) |
| Large protein (L) | 7 (L) | RNP complex component; homo dimerizes; binds to genomic and antigenomic RNA, VP35, and VP30; mRNA capping enzyme | Genome replication and mRNA transcription | (Yang et al., 2019) |
IFNA1, interferon alpha 1; IFNB1, interferon beta 1; IRF3, interferon regulatory factor 3; MX1, MX dynamin like GTPAse 1 (formerly MxA); NEDD4, NEDD4 E3 ubiquitin protein ligase; NPC1, NPC intracellular cholesterol transporter 1; PRKRA, protein activator of interferon induced protein kinase EIF2AK2 (formerly PACT); RNP, ribonucleoprotein; STAT1, signal transducer and activator of transcription 1; TSG101, tumor susceptibility 101; VP, virus protein
Genome organization and replication
The dianlovirus genome has the gene order 3′-NP-VP35-VP40-GP-VP30-VP24-L-5′ (Figure 1.Dianlovirus). The undetermined extragenic sequences at the extreme 3′-end (leader) and 5′-end (trailer) of the genome are assumed to be conserved and to be partially complementary. Genes are flanked by conserved transcriptional initiation and termination (polyadenylation) sites. Four dianlovirus genes overlap (Yang et al., 2019).
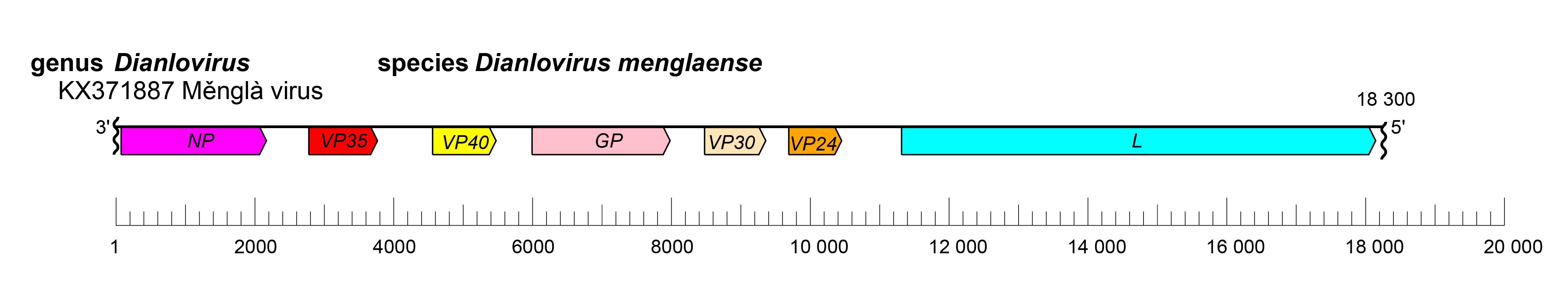 |
| Figure 1.Dianlovirus. Schematic representation of dianlovirus genome organization. Genome is drawn to scale. Wavy lines indicate incomplete genome ends. |
The replication strategy of dianloviruses remains to be studied but is assumed to be highly similar to that of orthomarburgviruses and reminiscent of that of cuevaviruses and orthoebolaviruses. Dianlovirions are assumed to associate with attachment factors at the plasma membrane that mediate infection by endocytosis. Dianlovirus GP1 and GP2 are assumed to mediate cell surface C-type lectin binding and subsequent low-pH-dependent fusion into endosomes, followed by GP1 and GP2 binding to the endosomal receptor NPC intracellular cholesterol transporter 1 (NPC1), which is also used by cuevaviruses, orthoebolaviruses, and orthomarburgviruses. Uncoating is presumed to occur in a manner analogous to that of other mononegaviruses. Dianlovirus transcription and genome replication likely take place in the cytoplasm and, in general, follow the models for other members of the Filoviridae family. Transcription starts at the conserved transcriptional initiation site, and polyadenylation occurs at a stretch of uridine residues within the transcriptional termination site. The 5′-terminal non-coding sequences favor hairpin like structures for all mRNAs. Replication involves the synthesis of full-length positive-sense copies (antigenomes). During infection, it is assumed that massive amounts of nucleocapsids accumulate intracellularly and form intracytoplasmic inclusion bodies. Virions are likely released via budding from plasma membranes (Figure 3.Filoviridae) (Yang et al., 2019).
Biology
Dianloviruses were discovered in 2019 by high-throughput sequencing of samples taken from an apparently healthy Rousettus sp. bat in China (Yang et al., 2017, Yang et al., 2019, Makenov et al., 2023).
Antigenicity
Initial studies indicate that MLAV is antigenically distinct from other filovirids, but more closely related to orthomarburgviruses than to orthoebolaviruses (Sherwood and Hayhurst 2019).
Species demarcation criteria
The genus currently includes only a single species.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Bat7633A47 | MT081488; MT081489; MT081490 | (Paskey et al., 2020) | |
| BtFiloYN2187 | KX371877 | (Yang et al., 2017) | |
| BtFiloYN2188 | KX371878 | (Yang et al., 2017) | |
| BtFiloYN2196 | KX371880 | (Yang et al., 2017) | |
| BtFiloYN2199 | KX371881 | (Yang et al., 2017) | |
| BtFiloYN2202 | KX371882 | (Yang et al., 2017) | |
| Vietnam-R.leschenaultii-119-2020 | OP653719* | (Makenov et al., 2023) | |
| Vietnam-R.leschenaultii-122-2020 | OP653720* | (Makenov et al., 2023) |
Virus names, the choice of exemplar isolates, and virus abbreviations, are not official ICTV designations.

