Family: Arenaviridae
Genus: Reptarenavirus
Distinguishing features
Reptarenaviruses infect snakes, and some cause boid inclusion body disease (BIBD) in snakes (Stenglein et al., 2012, Hetzel et al., 2013). Reptarenaviruses are notable for encoding a glycoprotein (GP) subunit GP2 that is more similar in structure to the glycoproteins of ebolaviruses (Mononegavirales: Filoviridae) than to those encoded by antennaviruses, hartmaniviruses, innmoviruses, and mammarenaviruses. In addition, the stable signal peptide (SSP), which remains associated with the GP complex in hartmaniviruses and mammarenaviruses, is lacking in the reptarenavirus GP complex (Koellhoffer et al., 2014).
Virion
Morphology
Virions are spherical or pleomorphic, 100–200 nm in dimeter, with dense lipid envelopes (Figure 1.Reptarenavirus). The virion surface layer is covered with club-shaped projections, 10 nm in length and 11–15 nm apart. These projections are made of trimeric spike structures of two virus-encoded membrane GP subunits (GP1 and GP2). Like the virions of mammarenaviruses, but unlike those of antennaviruses, innmoviruses, and hartmaniviruses, the virions of reptarenaviruses contain a zinc-binding protein (Z) layer under the membrane. In contrast to mammarenaviruses, no second layer exists beneath the membrane (Hetzel et al., 2013).
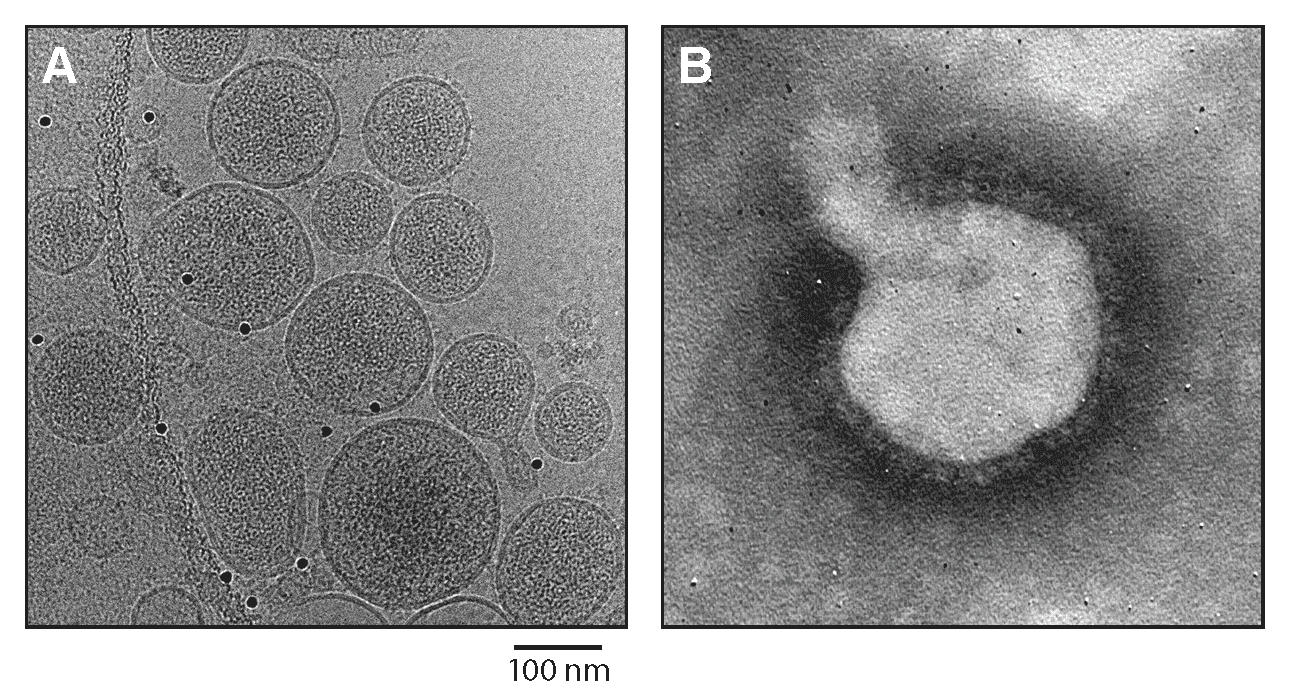 |
| Figure 1.Reptarenavirus. (A) Cryo-electron micrograph of University of Helsinki virus 1 (UHV1). Courtesy of Pasi Laurinmäki and Sarah Butcher, Cryo-EM Core Facility, Biocenter Finland, University of Helsinki, Finland. (B) Negative-stain electron micrograph of UHV1. Courtesy of Inki Luoto, University of Helsinki, Finland. |
Physicochemical and physical properties
Not reported
Nucleic acid
Reptarenaviruses have two ambisense single-stranded RNA segments that are encapsidated independently. The termini of the RNAs contain inverted complementary sequences encoding transcription and replication initiation signals (Stenglein et al., 2012, Hetzel et al., 2013, Hepojoki et al., 2018).
Proteins
Reptarenaviruses express four structural proteins. The most abundant structural protein in reptarenavirions is nucleoprotein (NP), which encapsidates the virus genomic segments. The least abundant protein is the large protein (L), which mediates virus genome replication and transcription. Z functions as a matrix protein. Unlike mammarenavirus Z, reptarenavirus Z does not possess an N-terminal glycine residue, typically associated with myristoylation for membrane anchoring. Instead, reptarenavirus Z has a predicted transmembrane domain located in the first 50 amino-acid residues that may serve a similar role. Reptarenavirus Z also does not contain late domain motifs that mediate viral budding. Instead, these motifs are found at the C terminus of the NP protein. Glycoprotein subunits GP1 and GP2 are derived by post-translational cleavage from an intracellular GPC. Reptarenaviruses do not produce SSP. The reptarenavirus GP complex is unique to the genus (Stenglein et al., 2012, Hetzel et al., 2013, Koellhoffer et al., 2014).
Lipids
Not reported
Carbohydrates
Not reported
Genome organization and replication
The small and large RNAs of reptarenaviruses encode two proteins in non-overlapping open reading frames (ORFs) of opposite polarities (ambisense coding arrangement) that are separated by non-coding intergenic regions (IGRs) (Figure 2.Reptarenavirus). The S RNA encodes NP in the virus genome-complementary sequence and the GPC in the virus genome-sense sequence. The L RNA encodes the L protein in the virus genome-complementary sequence and Z in the virus genome-sense sequence. The IGRs form one or more energetically stable stem–loop (hairpin) structures and function in structure-dependent transcription termination and in virion assembly and budding (Stenglein et al., 2012, Hetzel et al., 2013). In contrast to mammarenaviruses and hartmaniviruses, there is evidence that virus recombination events occur in snakes infected with reptarenaviruses (Stenglein et al., 2015).
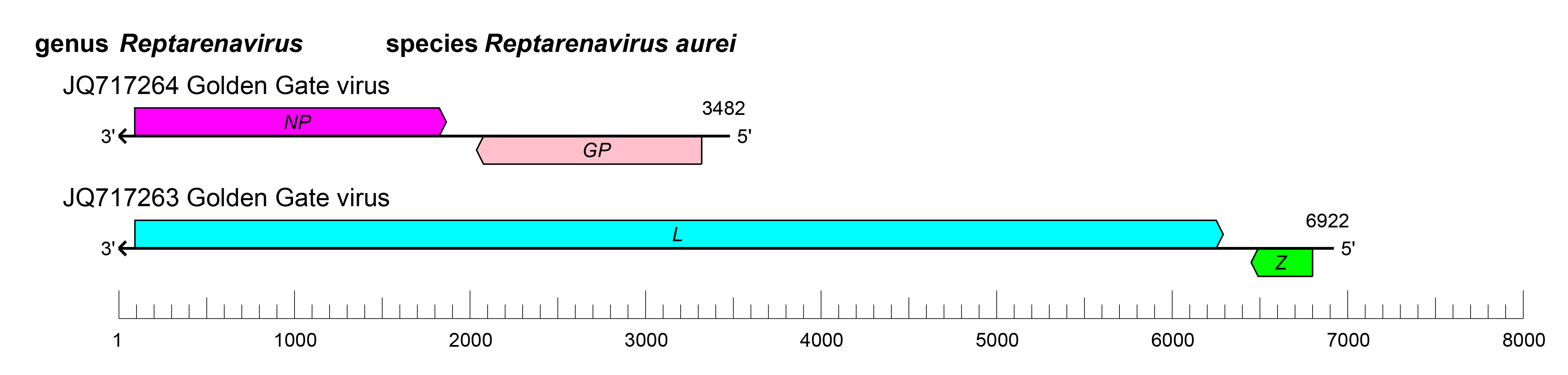 |
| Figure 2.Reptarenavirus. Schematic representation of reptarenavirus genome organization. The 5′- and 3′-ends of both segments are complementary at their termini, likely promoting the formation of circular ribonucleoprotein (RNP) complexes within the virion. GP, glycoprotein gene; L, large protein gene; NP, nucleoprotein gene; Z, zinc-binding protein gene. |
Biology
Some reptarenaviruses can cause acute or chronic BIBD in snakes belonging to the families Boidae and Pythonidae (Stenglein et al., 2012, Hetzel et al., 2013). In boas (Squamata: Boidae: Boa Linnaeus, 1758), disease outcome varies; affected animals either die within weeks or months or become asymptomatic (chronic) reptarenavirus carriers. In contrast, pythons generally develop severe fatal neurological symptoms within a few weeks. Virus replication sites also differ between snakes of these families. High virus loads may be detected in tissues (blood, liver, lungs, tonsils, spleen, kidneys, colon, trachea, brain, and skin) or excreta (feces and urine) of boas, whereas virus replication is limited to the central nervous system (CNS) tissues in pythons (Stenglein et al., 2017). Virus immunosuppression is thought to be a significant component of the disease. However, snakes do develop antibodies against reptarenaviruses (Hetzel et al., 2013, Korzyukov et al., 2016, Windbichler et al., 2019). In some cases, reptarenavirus infections are cleared without the host developing BIBD. The cytoplasmic inclusion bodies observed in BIBD are comprised of reptarenavirus NP and are located within mitochondria of infected cells (Baggio et al., 2021).
It remains unclear whether snakes are the natural host of reptarenaviruses or if they are infected incidentally. Historically, reptarenaviruses have been found in captive snakes, and only recently documented in wild boa constrictors in Costa Rica. Although horizontal and vertical transmission of reptarenaviruses has been reported in boa constrictors, the mechanism of transmission remains unclear. Several studies have identified multiple reptarenaviruses in the same snake, indicating that coinfections are common (Hepojoki et al., 2015, Stenglein et al., 2015, Keller et al., 2017). In experimental settings, reptarenaviruses can infect laboratory mice, causing mild to moderate lesions (without fatal outcome) in the liver, kidney, spleen, brain, and lungs (Abba et al., 2017).
Antigenicity
Antibodies against the mammarenaviruses lymphocytic choriomeningitis virus (LCMV) NP and Machupo virus (MACV) NP react weakly with the NP of the reptarenavirus University of Helsinki virus 1 (UHV-1). Human and rabbit anti-MACV sera also recognize UHV-1 NP (Hetzel et al., 2013). Systematic antigenicity studies for reptarenavirions have yet to be reported.
Species demarcation criteria
The parameters used to assign viruses to new species in the genus are:
- the virus shares less than 80% nucleotide sequence identity in the S RNA segment and less than 76% identity in the L RNA segment with other viruses;
- association of the virus with a distinct main host or a group of sympatric hosts;
- dispersion of the virus in a distinct defined geographical area; and/oror
- the virus shares less than 88% NP amino-acid sequence identity with other viruses (Radoshitzky et al., 2015).
Relationships within the genus
Phylogenetic relationships across the genus have been established from maximum-likelihood trees generated using full or partial sequences of NP and L proteins (Figure 3.Reptarenavirus).
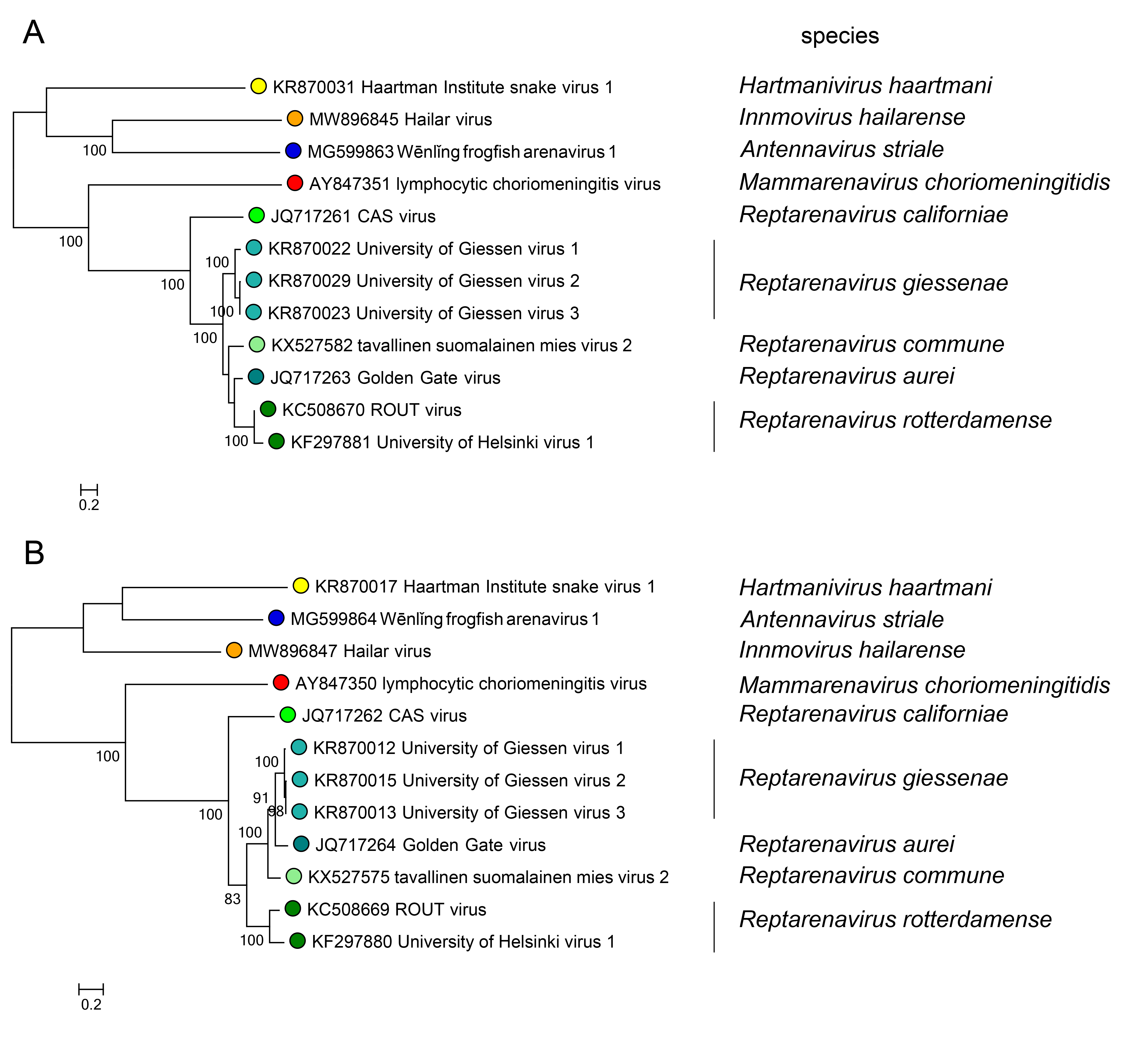 |
| Figure 3.Reptarenavirus. Maximum-likelihood phylogenetic trees inferred from MAFFT alignments (Katoh and Standley 2013) of the large (L) protein (A) and nucleoprotein (NP) (B) amino-acid sequences. The trees were generated by the IQ-TREE software v.1.6.12 (Trifinopoulos et al., 2016) using the best-fitting evolutionary model. Branch supports were calculated using the ultrafast bootstrap method (1,000 bootstraps). The mid-point rooted trees were visualized using FigTree (http://tree.bio.ed.ac.uk/). For NP and L protein, eight classified viruses (light green dots) were included. In both trees, representative viruses of the genera Mammarenavirus (red dot), Antennavirus (cyan dot), Hartmanivirus (dark green dot), and Innmovirus (yellow dot) are also included. |
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| aurora borealis virus 1 | S segment: KR870010*; L segment: KR870021 | ABV-1 | (Hepojoki et al., 2015) |
| aurora borealis virus 2 | S segment: KR870018 | ABV-2 | (Hepojoki et al., 2015) |
| aurora borealis virus 3 | L segment: KX527583 | ABV-3 | (Keller et al., 2017) |
| aurora borealis virus 4 | L segment: KX527592 | ABV-4 | (Keller et al., 2017) |
| bis spoeter virus | L segment: KX527598 | BSV-1 | (Keller et al., 2017) |
| boa Av DE1 | (Aqrawi et al., 2015) | ||
| boa Av DE2 (Aqrawi et al., 2015) | (Aqrawi et al., 2015) | ||
| boa Av DE3 | (Aqrawi et al., 2015) | ||
| boa Av DE4 | (Aqrawi et al., 2015) | ||
| Collierville virus | CVV | (Stenglein et al., 2012) | |
| corn snake reptarenavirus | S segment: KY072972 | (Hyndman et al., 2019) | |
| gruetzi mitenand virus 1 | L segment: KX527593* | GMV-1 | (Keller et al., 2017) |
| Hans Kompis virus 1 | L segment: KR870028 | HKV-1 | (Hepojoki et al., 2015) |
| hipoen jatkoon virus 1 | L segment: MH483048 | HJV-1 | (Hepojoki et al., 2018) |
| Kaltenbach virus 1 | L segment: KX527584 | KaBV-1 | (Keller et al., 2017) |
| keijut pohjoismaissa virus 1 | L segment: MH483047 | KePV-1 | (Hepojoki et al., 2018) |
| kiva uusi käärme virus 1 | L segment: MH483057 | KUKV-1 | (Hepojoki et al., 2018) |
| kuka mitä häh virus 1 | L segment: KX527588 | KMHV-1 | (Keller et al., 2017) |
| mistä näitä tulee virus 1 | L segment: MH483087 | MNTV-1 | (Hepojoki et al., 2018) |
| peilihimmeli vakooja virus 1 | L segment: MH483056 | PVaV-1 | (Hepojoki et al., 2018) |
| peto jauhoksi virus 1 | L segment: MH503956 | PJV-1 | (Hepojoki et al., 2018) |
| python Av DE1 | (Aqrawi et al., 2015) | ||
| rough scale python reptarenavirus | S segment: KY072969* | (Hyndman et al., 2019) | |
| S2-like virus | S segment: MH483056 | (Hepojoki et al., 2018) | |
| S5-like virus | S segment: KX527579 | (Keller et al., 2017) | |
| S7-like virus | S segment: MH483088 | (Hepojoki et al., 2018) | |
| S10-like virus | S segment: MH503957 | (Hepojoki et al., 2018) | |
| Stimson′s python 2 reptarenavirus | S segment: KY072971* | (Hyndman et al., 2019) | |
| Stimson′s python 5 reptarenavirus | S segment: KY072970* | (Hyndman et al., 2019) | |
| suri vanera virus 1 | L segment: KX527585 | SVaV-1 | (Keller et al., 2017) |
| suri vanera virus 2 | L segment: KX527587 | SVaV-2 | (Keller et al., 2017) |
| tavallinen suomalainen mies virus 1 | L segment: KX527595 | TSMV-1 | (Keller et al., 2017) |
| tavallinen suomalainen mies virus 2 | L segment: MH483050 | TSMV-2 | (Hepojoki et al., 2018) |
| University of Giessen virus 4 | S segment: KR870014 | UGV-4 | (Hepojoki et al., 2015) |
| University of Giessen virus S6-like | S segment: KX527578 | (Keller et al., 2017) | |
| University of Helsinki virus 3 | L segment: MH503952 | UHV-3 | (Hepojoki et al., 2018) |
| University of Helsinki virus 4 | L segment: KR870027 | UHV-4 | (Hepojoki et al., 2015) |
| UPM-MY 01 virus | L segment: KU198322* | (Abba et al., 2016) | |
| UPM-MY 02 virus | L segment: KU311007* | (Abba et al., 2016) | |
| UPM-MY 03 virus | L segment: KU311008* | (Abba et al., 2016) | |
| UPM-MY 04 virus | L segment: KU311009* | (Abba et al., 2016) |
Virus names and abbreviations are not official ICTV designations.
*Coding region sequence incomplete.

