Family: Arenaviridae
Genus: Antennavirus
Distinguishing features
Wēnlǐng frogfish arenavirus 1 (WlFAV1), Wēnlǐng frogfish arenavirus 2 (WlFAV2), salmon pescarenavirus 1 (SPAV1), and salmon pescarenavirus 2 (SPAV2) are the only known antennaviruses. All viruses have been found in fish (Shi et al., 2018, Mordecai et al., 2019). Notably, antennaviruses have genomes consisting of three genomic segments and likely do not encode a protein homologous with the zinc-binding protein (Z) of mammarenaviruses and reptarenaviruses.
Virion
Virions are unknown.
Physicochemical and physical properties
Not reported
Nucleic acid
Antennaviruses have one ambisense and two negative-sense RNA segments. The termini of the RNAs contain inverted complementary strands, likely encoding transcription and replication initiation signals (Shi et al., 2018).
Proteins
Based on sequence data only, antennaviruses likely express four structural proteins: nucleoprotein (NP), glycoprotein precursor (GPC), large (L) protein, and a protein of unknown function (Shi et al., 2018).
Genome organization and replication
The small (S) RNA of antennaviruses encodes NP; the medium (M) RNA encodes the GPC and an unknown protein; and the L RNA encodes the L protein, which contains an RNA-directed RNA polymerase (RdRP) domain (Figure 1.Antennavirus) (Shi et al., 2018).
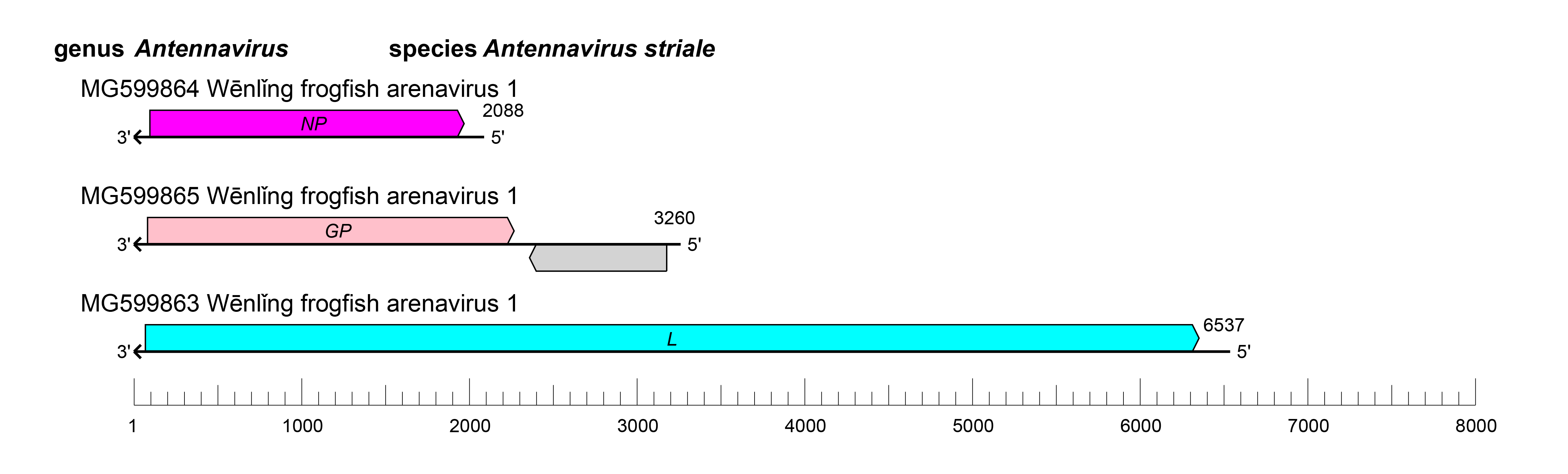 |
| Figure 1.Antennavirus. Schematic representation of antennavirus genome organization. The 5′- and 3′-ends of all segments (are complementary at their termini, likely promoting the formation of circular ribonucleoprotein (RNP) complexes within the virion. GP, glycoprotein gene; NP, nucleoprotein gene. |
Biology
Antennaviruses were discovered by high-throughput sequencing of samples from striated frogfish [Lophiiformes: Antennariidae: Antennarius striatus (G. Shaw, 1794)] captured by fishing trawlers in the East China Sea (Shi et al., 2018) and sockeye [Salmoniformes: Salmonidae: Oncorhynchus nerka (Walbaum, 1792)] and Chinook [Salmoniformes: Salmonidae: Oncorhynchus tshawytscha (Walbaum, 1792)] salmon sampled in the northeast Pacific (Mordecai et al., 2019). No antennavirus isolate has been documented.
Species demarcation criteria
The parameters used to assign a virus to a new species in the genus are (Radoshitzky et al., 2015):
- the virus shares less than 80% nucleotide sequence identity in the S RNA segment and less than 76% identity in the L RNA segment with other viruses;
- association of the virus with a distinct main host or a group of sympatric hosts;
- dispersion of the virus in a distinct defined geographical area;
- the virus shares less than 88% NP amino-acid sequence identity with other viruses(Radoshitzky et al., 2015).
Relationships within the genus
Phylogenetic relationships across the genus have been established from maximum-likelihood trees generated using complete sequences of NP and L proteins (Figure 2.Antennavirus).
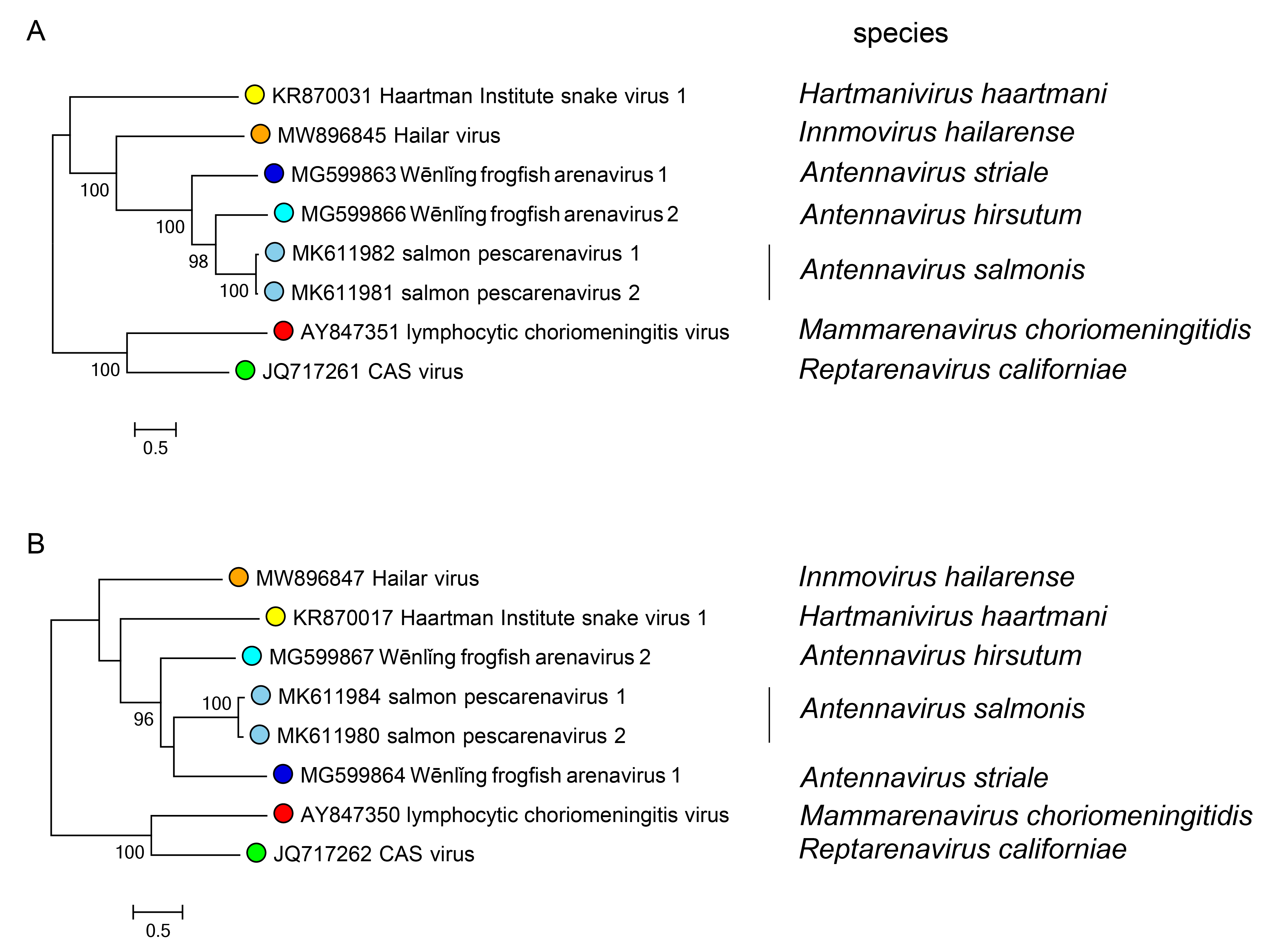 |
| Figure 2.Antennavirus. Maximum-likelihood phylogenetic trees inferred from MAFFT alignments (Katoh and Standley 2013) of the large (L) protein (A) and nucleoprotein (NP) (B) amino-acid sequences. The trees were generated by the IQ-TREE software v.1.6.12 (Trifinopoulos et al., 2016) using the best-fitting evolutionary model. Branch supports were calculated using the ultrafast bootstrap method (1,000 bootstraps). The mid-point rooted trees were visualized using FigTree (http://tree.bio.ed.ac.uk/). For NP and L protein, four classified viruses (cyan dots) were included. In both trees, representative viruses of the genera Mammarenavirus (red dot), Reptarenavirus (light green dot), Hartmanivirus (dark green dot), and Innmovirus (yellow dot) are also included. |
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| longfin eel arenavirus | (Grimwood et al., 2023) |
Virus names and abbreviations are not official ICTV designations.

