Family: Arenaviridae
Genus: Mammarenavirus
Distinguishing features
Mammarenaviruses typically infect muroid rodents. One mammarenavirus, Tacaribe virus (TCRV), has been found in phyllostomid bats and lone star ticks (Downs et al., 1963, Sayler et al., 2014), and recently discovered unclassified mammarenaviruses have been found in bats, hedgehogs, and pikas (Cui et al., 2023, Luo et al., 2023, Reuter et al., 2023). In contrast to antennaviruses and reptarenaviruses but similar to hartmaniviruses, mammarenaviruses encode a stable signal peptide (SSP), which remains associated with the glycoprotein (GP) complex (Buchmeier et al., 1987, Lenz et al., 2001, Eichler et al., 2003, Kunz et al., 2003, York et al., 2004, Bederka et al., 2014).
Virion
Morphology
Virions are spherical or pleomorphic, 50–200 nm in diameter, with dense lipid envelopes (Figure 1 Mammarenavirus). The virion surface layer is covered with club-shaped projections, 8–10 nm in length and 10 nm apart, with distinctive stalk and head regions. These projections are made of trimeric spike structures of two virus-encoded membrane GP subunits (GP1 and GP2). Isolated ribonucleoprotein (RNP) complexes are organized in “beads-on-a-string”-like structures. Internal granules within the virions are morphologically similar to host ribosomes (Dalton et al., 1968, Buchmeier 2002, Meyer et al., 2002, Charrel and de Lamballerie 2003, Jay et al., 2005, Neuman et al., 2005, Gonzalez et al., 2007, Li et al., 2016). Tomographic three-dimensional reconstructions of Lassa virus (LASV) particles and cryo-electron micrographic images of lymphocytic choriomeningitis virus (LCMV), Pichindé virus (PICHV), and TCRV particles have identified two interior layers beneath the inner leaflet of the virion lipid bilayer (Neuman et al., 2005, Li et al., 2016).
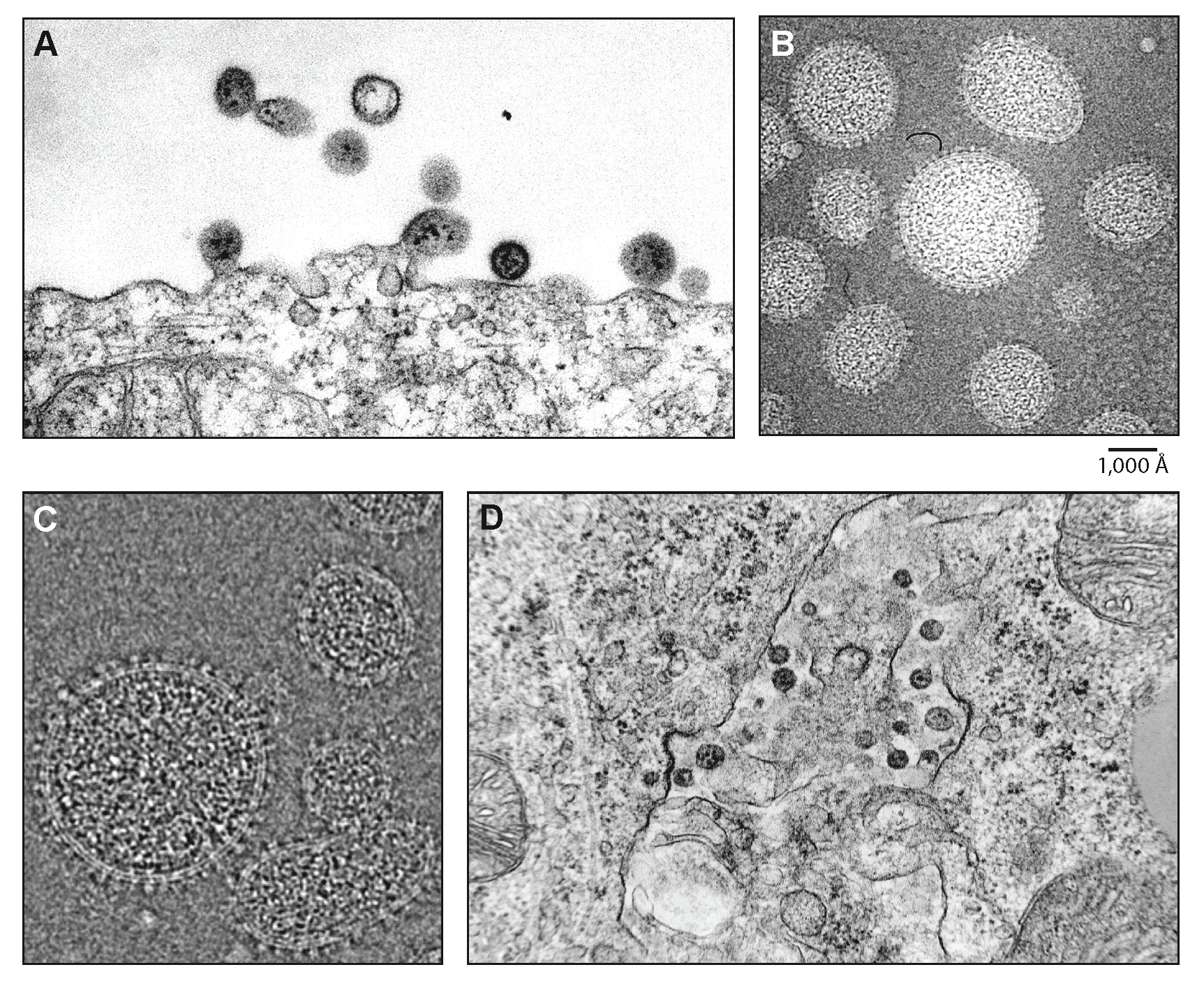 |
| Figure 1 Mammarenavirus. (A) Electron microscopic images of lymphocytic choriomeningitis virus (LCMV). The thin section shows several virions with internal inclusion bodies budding from the surface of an infected baby hamster kidney (BHK-21) cell. (B) Cryo-electron microscopic images of purified unstained LCMV particles frozen in vitreous ice. The bar indicates 100 nm. (C) Cryo-electron microscopy of frozen-hydrated Tacaribe virus (TCRV). The heterogeneous size and shape of these virions is typical of all of the viruses examined. (D) LCMV budding into intracellular spaces in the adrenal cortex of a guinea pig exposed 8 days earlier. (Courtesy R. Milligan, J. Burns, and M. Buchmeier). |
Physicochemical and physical properties
Virion molecular mass has not been determined. The sedimentation coefficient (S20,w) of virions is 325–500 S and virion buoyant density in sucrose, CsCl gradient centrifugation, and amidotrizoate compounds is about 1.17–1.18 g cm−3, 1.19–1.20 g cm−3, and 1.14 g cm−3, respectively. Virions are relatively unstable in vitro and are rapidly inactivated below pH 5.5 and above pH 8.5. Virus infectivity is inactivated at 56°C, by treatment with organic solvents or detergents, or with exposure to ultraviolet and gamma irradiation (Pfau 1965, Rawls and Buchmeier 1975, Elliott et al., 1982, Mitchell and McCormick 1984).
Nucleic acid
Mammarenaviruses have two ambisense single-stranded RNA segments that are encapsidated independently. The termini of the RNAs contain inverted complementary sequences encoding transcription and replication initiation signals. The 3′-ends of the untranslated regions (UTRs) of the genome and antigenome segments contain the genomic and antigenomic promoters, respectively, that direct RNA replication and gene transcription (Perez et al., 2003, Hass et al., 2006). No poly(A) tracts are present at the 3′-end. The 5′- and 3′-ends of the large (L) and small (S) RNA segments contain conserved reverse-complementary sequences of 19–30 nucleotides at each extremity (Auperin et al., 1982a, Auperin et al., 1982b). These termini are predicted to form panhandle structures through base pairing (Young and Howard 1983, Salvato et al., 1989, Harnish et al., 1993). Although the genomic RNAs are thought to be present in virions in the form of circular nucleocapsids, the genomic RNA is not covalently closed. Virus RNAs are not present in equimolar amounts in virions, apparently due to the packaging of multiple RNA molecules per virion. For example, more than one S RNA molecule might be packaged per virion (Romanowski and Bishop 1983).
Proteins
Mammarenaviruses express four structural proteins (Table 1 Mammarenavirus). The most abundant structural protein in mammarenavirions is nucleoprotein (NP), which encapsidates the virus genomic segments. The least abundant protein is the L protein, which mediates virus genome replication and transcription. The zinc-binding protein (Z) functions as a matrix protein. Unlike reptarenavirus Z, mammarenavirus Z (with the exception of TCRV Z) possesses C-terminal canonical late domain motifs involved in virus budding and an N-terminal glycine residue, typically associated with myristoylation for membrane anchoring. The preproprotein GP precursor (GPC; 75–76 kD) is cleaved by the signal peptidase to generate the SSP and a precursor protein that is processed by the cellular protease subtilisin kexin isozyme-1/site 1 protease (SKI-1/S1P) to generate the mature virion surface glycoproteins (GP1 and GP2).
Table 1 Mammarenavirus. Location and functions of mammarenavirus structural proteins.
| Protein | Location, mass, and function |
| Nucleoprotein (NP) | Structural virion protein (60–68 kD). Component of the ribonucleoprotein (RNP) inside virions. Oligomerizes and encapsidates virus genomic and antigenomic segments. Functions as an exoribonuclease and serves as an interferon antagonist. Interacts with Z (Buchmeier 2002, Martínez-Sobrido et al., 2006, Brunotte et al., 2011, Hastie et al., 2011a, Hastie et al., 2011b, West et al., 2014). |
| Glycoprotein (GP) | Structural virion glycoprotein (50–60 kD). Produced via proteolytic cleavage from the GP precursor (GPC; about 70–80 kD). Cleavage produces heterotrimers consisting of GP1, GP2, and stable signal peptide (SSP). Inserts into virion membranes as a tripartite heterotrimeric GP complex. As a class I fusion machine, GP mediates cell-surface and internal receptor binding (via GP1), virion-cell membrane fusion and, thereby, virion cell entry (via GP2) (Buchmeier et al., 1987, Lenz et al., 2001, Eichler et al., 2003, Kunz et al., 2003, York et al., 2004, Bederka et al., 2014, Hastie et al., 2017). |
| Large (L) protein | Structural virion protein (250–450 kD). Component of the RNP inside virions. Via its RNA-directed RNA polymerase domain, mediates transcription and replication of mammarenavirus RNA segments. Mediates cap-snatching for virus messenger RNA capping (Kranzusch et al., 2010, Reguera et al., 2016, Peng et al., 2020, Kouba et al., 2021). |
| Zinc-binding (Z) protein | Structural virion protein (10–14 kD). As a virion matrix protein, Z self-associates, polymerizes at membranes, mediates virion assembly and budding, interacts with NP and the GPC, and negatively regulates transcription. Z also serves as an interferon antagonist (Salvato and Shimomaye 1989, Perez et al., 2003, Perez et al., 2004, Strecker et al., 2006, Capul et al., 2007, Urata et al., 2009, Fehling et al., 2012, Hastie et al., 2016, Shao et al., 2018). |
Lipids
Lipids represent about 20% of virion dry weight and are similar in composition to those of the host plasma membrane. Z and SSP of mammarenaviruses are myristoylated at glycine residue 2. Myristoylation is critical for Z to bind to lipid membranes and thus for virion budding, whereas myristoylation of SSP is critical for membrane fusion (Perez et al., 2004, York et al., 2004).
Carbohydrates
Carbohydrates in the form of complex glycans are present on GP1 and GP2 and represent up to 8% of virion dry weight (Grutadauria et al., 1999, Bonhomme et al., 2011). These carbohydrates serve as a shield for virions from host antibodies (Sommerstein et al., 2015, Watanabe et al., 2018).
Genome organization and replication
The S and L RNAs of mammarenaviruses each encode two proteins in non-overlapping open reading frames (ORFs) of opposite polarities (ambisense coding arrangement) that are separated by non-coding intergenic regions (IGRs) (Figure 2 Mammarenavirus). The S RNA encodes NP in the virus genome-complementary sequence and the GPC in the virus genome-sense sequence. The L RNA encodes the L protein in the virus genome-complementary sequence and Z in the virus genome-sense end sequence (Salvato and Shimomaye 1989). The IGRs form one or more energetically stable stem–loop (hairpin) structures (Auperin et al., 1984a, Auperin et al., 1984b, Wilson and Clegg 1991) and function in structure-dependent transcription termination (Meyer and Southern 1993, Meyer and Southern 1994, Tortorici et al., 2001) and in virion assembly and budding (Pinschewer et al., 2005) (Figure 2.Mammarenavirus).
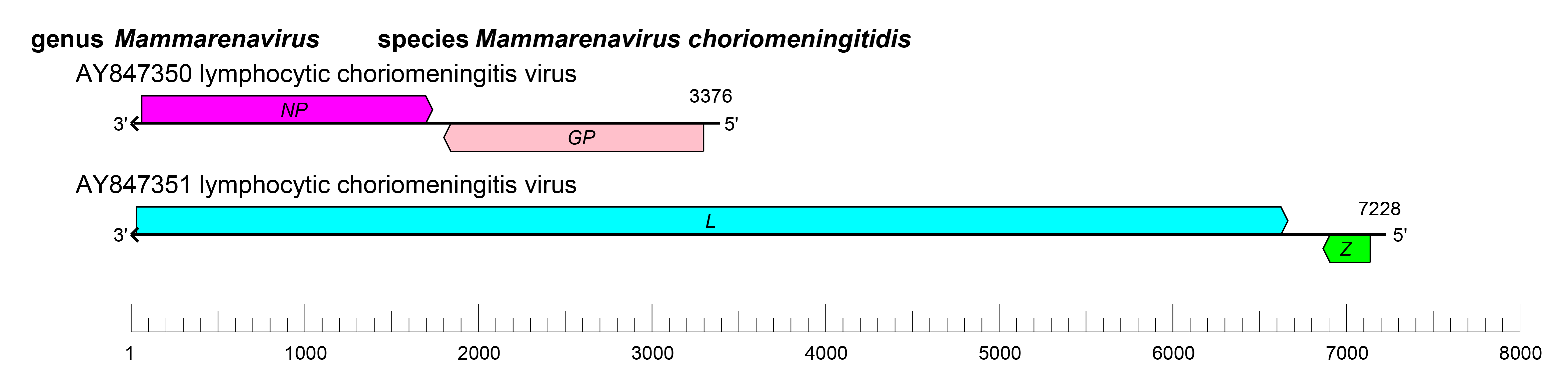 |
| Figure 2 Mammarenavirus. Schematic representation of the genome organization of lymphocytic choriomeningitis virus (LCMV). The 5′- and 3′-ends of both segments (small [S] and large [L]) are complementary at their termini, likely promoting the formation of circular ribonucleoprotein (RNP) complexes within the virion. GP, glycoprotein gene; L, large protein gene; NP, nucleoprotein gene; Z, zinc-binding protein gene. |
Mammarenavirus infection begins with virion attachment to cell-surface receptors (e.g., dystroglycan 1 [DAG1] or transferrin receptor [TFRC]) and entry via the endosomal route (Glushakova and Lukashevich 1989, Borrow and Oldstone 1994, Cao et al., 1998, Martinez et al., 2007, Radoshitzky et al., 2007, Vela et al., 2007, Raaben et al., 2017) (Figure 3.Arenaviridae). pH-dependent fusion with late endosomes releases the virion RNP complex into the cytoplasm. In the case of LASV and LCMV, entry involves a pH-dependent switch from DAG1 to an intracellular receptor, lysosomal associated membrane protein 1 (LAMP1) or the CD164 molecule, respectively (Jae et al., 2014, Bakkers et al., 2022). Likewise, completion of LUJV cell entry involves a switch from the viral cell surface receptor neuropilin (NRP)-2 to the intracellular entry factor tetraspanning protein CD63 (Raaben et al., 2017).
The virus RNP directs both RNA genome replication and gene transcription (Meyer et al., 2002). During replication, L protein reads through the IGR transcription-termination signal and generates uncapped antigenomic and genomic RNAs (Leung et al., 1977). Because these RNAs contain a single non-templated G at the 5′-ends (Garcin and Kolakofsky 1990, Raju et al., 1990), replication initiation might involve a slippage mechanism of L protein on the nascent RNA (Garcin and Kolakofsky 1992). Because of the ambisense coding arrangement, only mRNAs encoding NP or L protein can be synthesized from genomic RNAs. Transcription of mRNAs encoding GPC or Z occurs only after the first round of virus replication, during which S and L antigenomes are produced.
Mammarenavirus proteins are synthesized from subgenomic capped mRNAs, which are not polyadenylated (Singh et al., 1987, Southern et al., 1987, Meyer and Southern 1993). The 5′-ends of virus mRNAs contain several non-templated bases downstream of the 5′-cap structure, suggesting that mammarenaviruses use a cap-snatching mechanism similar to that used by other viruses of the negarnaviricot subphylum Polyploviricotina (Garcin and Kolakofsky 1990, Raju et al., 1990, Meyer and Southern 1993). Cap-snatching requires an endonuclease present in the N-terminal part of L protein that cleaves cellular mRNAs to generate a cap leader that is subsequently used to prime transcription (Kranzusch et al., 2010, Peng et al., 2020). The 3′-ends of the mRNAs have been mapped to locations in the IGRs.
The mammarenavirus GPC polyprotein matures in the lumen of the endoplasmic reticulum, where its SSP is co-translationally cleaved and GPC is extensively N-glycosylated. GPC is thought to oligomerize prior to proteolytic processing by SKI-1/S1P. Proteolytic maturation of GPC into GP1 and GP2 as well as trafficking of GP1 and GP2 from the endoplasmic reticulum to the cell surface is dependent on the SSP. Virion budding occurs from the cellular plasma membrane, thereby providing the virion′s envelope. Ribosome-like structures are observed within virions, but the origin and composition of these structures have not been elucidated (Dalton et al., 1968, Perez et al., 2003, Strecker et al., 2003, Eichler et al., 2004). Incorporation of these structures does not seem to be required for mammarenavirus replication and infectivity (Leung and Rawls 1977).
During mammarenavirus infection, intratypic reassortant progeny can be formed, including diploid (or multiploid) types with respect to the genomic RNA segments. Evidence for reassortment between viruses of different species (e.g., LASV and Mopeia virus [MPOV]) has also been obtained (Lukashevich 1992).
Biology
The reservoir hosts of almost all mammarenaviruses are rodents of the superfamily Muroidea (Bowen et al., 1997, Hugot et al., 2001). LCMV is found in the house mouse and has a worldwide distribution, whereas mammarenaviruses in Africa are found mainly in rodents belonging to the Mastomys and Praomys genera (family Muridae, subfamily Murinae). Mammarenaviruses in North and South America are mostly found in rodents belonging to the murid subfamily Cricetidae. An exception is TCRV, which was isolated from Jamaican fruit-eating bats (Chiroptera: Phyllostomidae: Artibeus jamaicensis Leach, 1821) and great fruit-eating bats (Artibeus lituratus Olfers, 1818) in the Caribbean (Downs et al., 1963) and from lone star ticks (Ixodida: Ixodidae: Amblyomma americanum (Linnaeus, 1758)) in Florida (Sayler et al., 2014). Some unclassified mammarenaviruses were discovered in bats (Bentim Góes et al., 2022), dipodoid rodents (jerboas) (Wu et al., 2018), hedgehogs (Reuter et al., 2023), and pikas (Cui et al., 2023, Luo et al., 2023).The geographic range of mammarenaviruses is generally much more restricted than that of their cognate rodent reservoir hosts (Salazar-Bravo et al., 2002a, Salazar-Bravo et al., 2002b, Gonzalez et al., 2007).
Typically, mammarenaviruses establish persistent, frequently asymptomatic infections characterized by chronic viremia and viruria in their reservoir hosts (Bowen et al., 1997, Hugot et al., 2001). It is suspected that chronic infections are caused by suppressed host immunity. The chronic carrier state in rodents usually results from vertical transmission (exposure to infectious virus early in ontogeny) (Webb et al., 1975, Mills et al., 1992).
Most mammarenaviruses do not normally infect mammals other than their primary reservoir hosts. However, mammarenaviruses have the ability to cross species barriers. Humans become infected via direct contact with infected muroid rodents or their droppings or urine; ingestion of contaminated food; or inhalation of aerosolized droplets from contaminated rodent excreta, secreta, or body parts (sometimes caught in mechanical harvesters) (Charrel and de Lamballerie 2003). Handling and consumption of infected rodents can be another route of virus transmission (Keenlyside et al., 1983). Person-to-person transmission of mammarenaviruses occurs by nosocomial routes and is rare but possible through direct contact with body fluids or excreta of infected patients (Johnson 1965, Fisher-Hoch et al., 1995, Paweska et al., 2009).
Human diseases caused by mammarenaviruses include Lassa fever (caused by LASV) in Western Africa. Lujo virus (LUJV) has caused a small but severe disease outbreak in Southern Africa. Junín virus (JUNV) causes Argentinian hemorrhagic fever in an increasingly large area of Argentina. Machupo virus (MACV) has caused isolated outbreaks of Bolivian hemorrhagic fever in Bolivia. Guanarito virus (GTOV) is the etiologic agent of Venezuelan hemorrhagic fever in Venezuela. Sabiá virus (SBAV) and Chapare virus (CHAPV) have been isolated from human cases with fatal outcomes in Brazil and Bolivia, respectively (Radoshitzky et al., 2018).
Cases of South American mammarenavirus infection have predominantly occurred among male agricultural workers who have come into contact with infected rodents during harvest season when rodent populations are active (Radoshitzky et al., 2018). Human infection with LCMV may occur in rural and urban areas with high rodent populations and has been acquired from pet hamsters (Hirsch et al., 1974). Organ transplants from LCMV-infected individuals have resulted in at least 10 human deaths in the United States since 1998 (MacNeil et al., 2012). Severe laboratory-acquired infections have occurred with Flexal virus (FLEV), JUNV, LASV, LCMV, MACV, and SBAV (Milzer and Levinson 1942, Rugiero et al., 1962, Leifer et al., 1970, Vasconcelos et al., 1993, Gaidamovich et al., 2000). Mild and asymptomatic human infections with PICHV have also been reported (Buchmeier et al., 1974, Gonzalez et al., 2007).
LCMV acquired from house mice has also caused a fatal hepatitis in various captive callitrichid primates, specifically marmosets and tamarins (Montali et al., 1993). Both virus and host factors contribute to the outcome of experimental mammarenavirus infection of laboratory animals, including:
- rodents such as big lauchas (Calomys callosus Rengger, 1830), drylands lauchas (Calomys musculinus (Thomas, 1913)), Chinese hamsters (Cricetulus griseus Milne-Edwards, 1867), golden hamsters (Mesocricetus auratus Waterhouse, 1839), guinea pigs (Cavia porcellus (Linnaeus, 1758)), laboratory mice, Molina′s akodonts (Akodon molinae Contreras, 1968), and Natal mastomys (Mastomys natalensis Smith 1834) (Justines and Johnson 1969, Terrell et al., 1973, Walker et al., 1975, Buchmeier and Rawls 1977, Lampuri et al., 1982, Лукашевич et al., 1985, Carballal et al., 1986);
- Jamaican fruit-eating bats (Cogswell-Hawkinson et al., 2012); and
- nonhuman primates such as common marmosets (Callithrix jacchus (Linnaeus, 1758)), crab-eating macaques (Macaca fascicularis Raffles, 1821), grivets (Chlorocebus aethiops (Linnaeus, 1758)), hamadryas baboons (Papio hamadryas (Linnaeus, 1758)), rhesus monkeys (Macaca mulatta (Zimmermann, 1780)), squirrel monkeys (Saimiri sciureus (Linnaeus, 1758)), and tufted capuchins (Cebus apella (Linnaeus, 1758)) (Wagner et al., 1977, Peters et al., 1987, Евсеев et al., 1991, Vela 2012, Golden et al., 2015).
In general, South American mammarenaviruses are pathogenic for suckling but not weaned laboratory mice, whereas the situation is reversed for African mammarenaviruses, including LCMV and LASV (Golden et al., 2015).
A diverse range of mammalian cell lines are susceptible to mammarenavirus infection in vitro.
Mammarenaviruses primarily infect cells of the myeloid and reticuloendothelial lineages but also hepatocytes, lymphocytes, and other cells (Johnson 1965, Maiztegui 1975, González et al., 1980, Hensley et al., 2011). Receptors mediating host cell entry are mainly DAG1 for some African mammarenaviruses (e.g., some strains of LCMV and LASV) and some South American mammarenaviruses (Cao et al., 1998, Spiropoulou et al., 2002), neuropilin-2 (NRP2) and CD63 for LUJV (Raaben et al., 2017, Saito et al., 2021), and TFRC for other South American mammarenaviruses (e.g., GTOV, JUNV, and MACV) (Radoshitzky et al., 2007). However, virus entry by some strains has been observed in the absence of or in addition to these receptors using alternative factors, such as AXL receptor tyrosine kinase(AXL), calcium voltage-gated channel subunit alpha1 S (CACNA1S), C-type lectin domain family 4 member G (CLEC4G), CD209, hepatitis A virus cellular receptor (HAVCR1), or TYRO3 protein tyrosine kinase (TYRO3) (Shimojima et al., 2012, Brouillette et al., 2018, Sarute and Ross 2020).
Antigenicity
Antigenic cross-reactions based on cross-protection, neutralization of infectivity, complement-fixation, and indirect immunofluorescence tests have been widely reported and involve determinants located primarily on NP and GP (Rowe et al., 1970b, Gajdamovič et al., 1975, Wolff et al., 1978, Buchmeier et al., 1980, Damonte et al., 1986, Sanchez et al., 1989). In addition, antibodies against the mammarenaviruses LCMV and MACV react weakly with the NP of the reptarenavirus University of Helsinki virus 1 (UHV1). Human and rabbit anti-MACV sera also recognize UHV1 NP (Hetzel et al., 2013). GP1 of mammarenaviruses is the major virus neutralization and protective antigen. Antibodies to NP and, variably, to other virus proteins are also induced by infection but generally do not neutralize infectivity in vitro. Major complement-fixing antigens are associated with NP. Furthermore, NP and GP of LCMV and LASV contain cytotoxic T-cell epitopes.
Species demarcation criteria
The parameters used to assign a virus to a new species in the genus are:
- the virus shares less than 80% nucleotide sequence identity in the S RNA segment and less than 76% identity in the L RNA segment with other viruses;
- association of the virus with a distinct main host or a group of sympatric hosts;
- dispersion of the virus in a distinct defined geographical area;
- association (or lack thereof) with human disease;
- the virus shares less than 88% NP amino-acid sequence identity with other viruses (Radoshitzky et al., 2015).
Relationships within the genus
Phylogenetic relationships across the genus have been established from maximum-likelihood trees generated using full or partial sequences of NP and L proteins (Figure 3 Mammarenavirus).
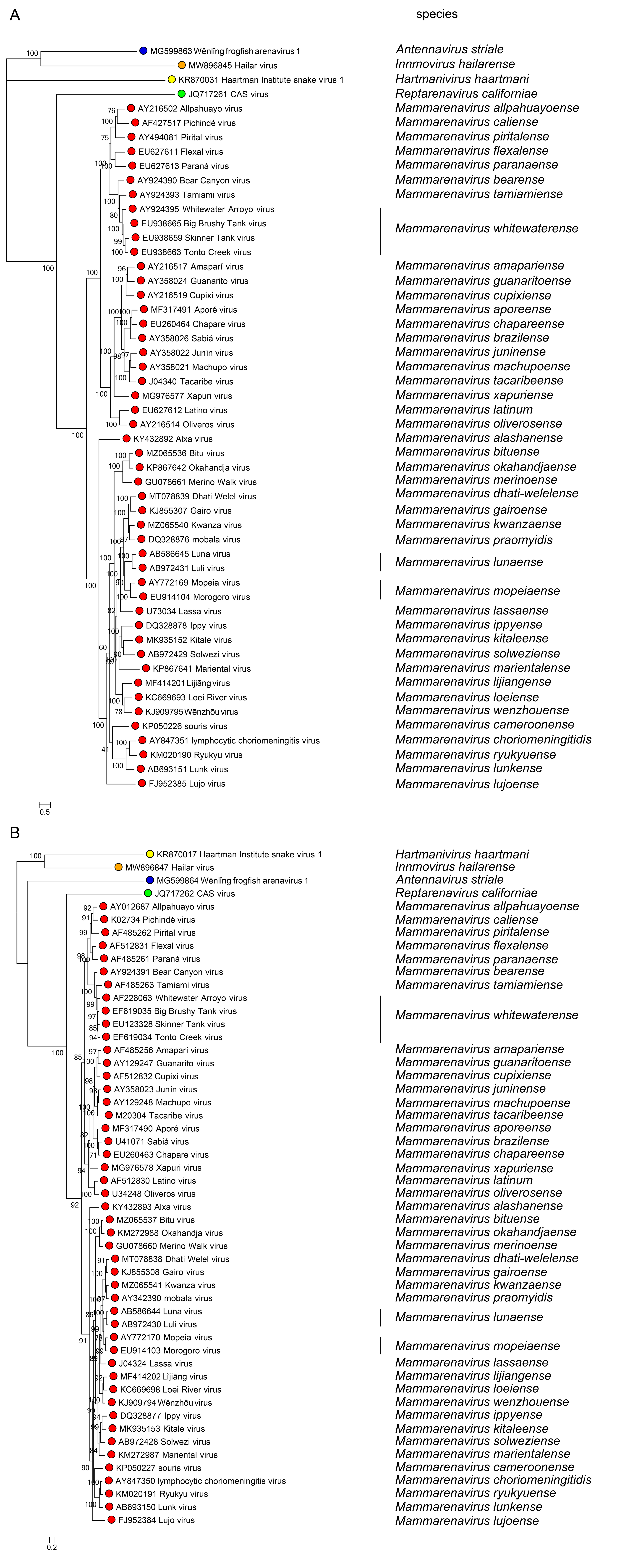 |
| Figure 3 Mammarenavirus. Maximum-likelihood phylogenetic trees inferred from MAFFT alignments (Katoh and Standley 2013) of the large (L) protein (A) and nucleoprotein (NP) (B) amino-acid sequences. The trees were generated by the IQ-TREE software v.1.6.12 (Trifinopoulos et al., 2016) using the best-fitting evolutionary model. Branch supports were calculated using the ultrafast bootstrap method (1,000 bootstraps). The mid-point rooted trees were visualized using FigTree (http://tree.bio.ed.ac.uk/). For NP and L protein, 48 classified viruses (red dots) were included. In both trees, representative viruses of the genera Antennavirus (cyan dot), Reptarenavirus (light green dot), Hartmanivirus (dark green dot), and Innmovirus (yellow dot) are also included. |
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | |
| Aershan mammarenavirus | L segment: ON012176* | ||
| arenavirus 96010025 | S segment: EU486820 | ||
| arenavirus DX1401 | S segment: KJ144198*; L segment: KJ144197* | ||
| arenavirus Mus/TZ22285/TZA/2008 | L segment: GU182413* | ||
| arenavirus sp. 9408800235/ZAF | S segment: FJ383127* | ||
| Black Mesa virus | S segment: FJ032026; S segment: FJ032027; L: EU938670* | ||
| Bobomene virus | S segment: KF926408* | ||
| Conseil virus | (Geoghegan et al., 2021) | ||
| Gbagroube virus | S segment: GU830848; L segment: GU830849* | (Coulibaly-N'Golo et al., 2011) | |
| jerboa arenavirus | L segment: MF642354* | JEAreV | (Wu et al., 2018) |
| Jirandogo virus | S segment: JX845169; L segment: JX845167* | (Kronmann et al., 2013) | |
| Kodoko virus | S segment: EF189586*; L segment: EF179864* | KODV | (Lecompte et al., 2007) |
| Menékré virus | S segment: GU830857*; L segment: GU830858* | (Coulibaly-N'Golo et al., 2011) | |
| Middle Pease River virus | S segment: JX657695* | MPRV | (Cajimat et al., 2013) |
| Natorduori virus | S segment: JX845170*; L segment: JX845168* | (Kronmann et al., 2013) | |
| North American arenavirus 96010024 | S segment: EU123331 | (Cajimat et al., 2013) | |
| North American arenavirus 96010151 | S segment: EU123330 | (Cajimat et al., 2013) | |
| North American arenavirus D1240007 | S segment: EU123329 | (Cajimat et al., 2013) | |
| Ocozocoautla de Espinosa virus | S segment: JN897398 | OCEV | (Cajimat et al., 2012) |
| Omdraaivlei virus | S segment: KF926410* | ||
| Orogrande virus | S segment: EU910959; L segment: EU938669* | ||
| Palo Verde virus | PVEV | (Milazzo et al., 2015) | |
| Patawa virus | S segment: KJ668824*; L segment: KJ668825* | (Lavergne et al., 2015) | |
| Pinhal virus | S segment: EU280547* | ||
| Pistillo virus | (Wildy 1971) | ||
| Real de Catorce virus | L segment: JF430461* | (Cajimat et al., 2011) | |
| Serodino virus | (Shedroff et al., 2023) | ||
| Witsand virus | S segment: KF926412* |
Virus names and abbreviations are not official ICTV designations.
*Coding region sequence are incomplete.

