Family: Arenaviridae
Sheli R. Radoshitzky, Michael J. Buchmeier, Rémi N. Charrel, Jean-Paul J. Gonzalez, Stephan Günther, Jussi Hepojoki, Jens H. Kuhn, Igor S. Lukashevich, Víctor Romanowski, Maria S. Salvato, Manuela Sironi, Mark D. Stenglein, and Juan Carlos de la Torre
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Juan Carlos de la Torre (juanct@scripps.edu)
Edited by: Jens H. Kuhn, Stuart G. Siddell, and Peter J. Walker
Posted: May 2019, updated September 2020, August 2023, May 2024
PDF: ICTV_Arenaviridae.pdf (2020 version)
Summary
Arenaviridae is a family for ambisense RNA viruses with genomes of about 10.5 kb (Table 1 Arenaviridae). The family includes five genera (Antennavirus, Hartmanivirus, Innmovirus, Mammarenavirus, and Reptarenavirus). These viruses infect fish (antennaviruses), snakes (hartmaniviruses and reptarenaviruses), mammals (mammarenaviruses), and unknown hosts (innmoviruses). Some reptarenaviruses cause boid inclusion body disease (BIBD) in captive snakes, whereas some mammarenaviruses can infect humans and other primates, causing mild, severe, and sometimes-fatal diseases. The genomes of arenavirids produce enveloped virions containing two or three single-stranded RNA segments with open reading frames (ORFs) that encode nucleoproteins (NPs), glycoproteins (GPs), and large (L) proteins containing RNA-directed RNA polymerase (RdRP) domains; some arenavirids encode zinc-binding proteins (Z).
Table 1 Arenaviridae. Characteristics of members of the family Arenaviridae.
| Characteristic | Description* |
| Example | lymphocytic choriomeningitis virus Armstrong 53b [S RNA segment: AY847350; L RNA segment: AY847351], species Mammarenavirus choriomeningitidis |
| Virion | Enveloped, pleomorphic virions 40–200 nm in diameter with trimeric surface spikes |
| Genome | Two or three single-stranded RNA molecules (segments): small (S), medium (M), and large (L), usually with ambisense coding arrangement |
| Replication | Ribonucleoprotein (RNP) complexes are generated that contain anti-genomic RNA serving as coding templates for synthesis of genomic RNA. |
| Translation | Proteins are produced from capped and non-polyadenylated mRNAs. The 5′ cap structure is obtained via cap-snatching from cellular mRNAs. |
| Host range | Fish (antennaviruses), mammals (mammarenaviruses), and reptiles (hartmaniviruses and reptarenaviruses), but potentially also ticks |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Polyploviricotina, class Bunyaviricetes, order Hareavirales; the family includes five genera and 71 species. |
* mostly based on experiments with mammalian arenavirids
Viruses assigned to each of the five genera form a monophyletic clade based on phylogenetic analysis of L protein/RdRP and NP sequences. Viruses from all five genera share one or more of the following characteristics: (i) enveloped spherical or pleomorphic virions; (ii) segmented single-stranded, ambisense RNA genome without polyadenylated tracts at the 3′-end; (iii) genomic sequence complementarity at the 5′- and 3′-ends; (iv) nucleotide sequences that could form one or more hairpin configurations within non-coding intergenic regions (IGRs) of genomic and antigenomic segments; (v) capped but not polyadenylated virus mRNAs; and (vi) induction of a persistent and frequently asymptomatic infection in reservoir hosts, in which chronic viremia and/or viruria may occur (Radoshitzky et al., 2015).
Piscine hosts
Genus Antennavirus. This genus includes four species for viruses discovered in actinopterygian fish. Antennavirusgenomes consist of three, rather than two, genomic segments and appear not to encode the Z protein of mammarenaviruses and reptarenaviruses.
Reptilian hosts
Genus Hartmanivirus. This genus includes eight species for 11 viruses discovered in captive snakes with BIBD. Hartmanivirus genomes lack a gene encoding the Z protein of mammarenaviruses and reptarenaviruses.
Genus Reptarenavirus. This genus includes five species for eight viruses discovered in captive snakes, some of which showed signs of BIBD. Reptarenaviruses are notable for their transmembrane surface glycoproteins (GP2), which are more closely related to those of ebolaviruses (Mononegavirales: Filoviridae) than to those of antennaviruses, hartmaniviruses, innmoviruses, mammarenaviruses, or other bunyavirals. Reptarenaviruses are also unusual in that the same host often harbors multiple distinct small (S) and large (L) segments, suggesting a high frequency of reptarenavirus co-infections with distinct viruses or the formation of viruses with the same L but distinct S segments.
Mammalian hosts
Genus Mammarenavirus. The genus includes 53 species for viruses detected in rodent hosts, apart from Tacaribe virus (TCRV), which has been found only in phyllostomid bats and ixodid lone star ticks. Mammarenavirus infections of their natural rodent hosts are generally asymptomatic. In humans, some mammarenaviruses, such as Western African Lassa virus (LASV) or several viruses of South American origin, can cause severe and often-fatal diseases with hemorrhagic manifestations. Lymphocytic choriomeningitis virus (LCMV), a typical mammalian arenavirus, may cause disease in humans and poses a serious threat to immunocompromised individuals.
Unknown hosts
Genus Innmovirus. This recently established genus includes a single species for a virus discovered in river sediment samples. Innmoviruses have genomes consisting of three, rather than two, genomic segments and appear not to encode a homologue of the Z protein of mammarenaviruses and reptarenaviruses.
Virion
Morphology
Virions are spherical or pleomorphic in shape, 40–200 nm in diameter, with dense lipid envelopes (Figure 1 Arenaviridae). The virion surface layer is covered with club-shaped projections with distinctive stalk and head regions. These projections consist of trimeric spike structures of two virus-encoded membrane GP subunits (GP1 and GP2) and, in some arenavirids, a third component (stable signal peptide [SSP]). Isolated ribonucleoprotein (RNP) complexes are organized into “beads-on-a-string”-like structures (Buchmeier 2002, Meyer et al., 2002, Charrel and de Lamballerie 2003, Jay et al., 2005, Neuman et al., 2005, Hetzel et al., 2013, Li et al., 2016, Hepojoki et al., 2018, Radoshitzky et al., 2020).
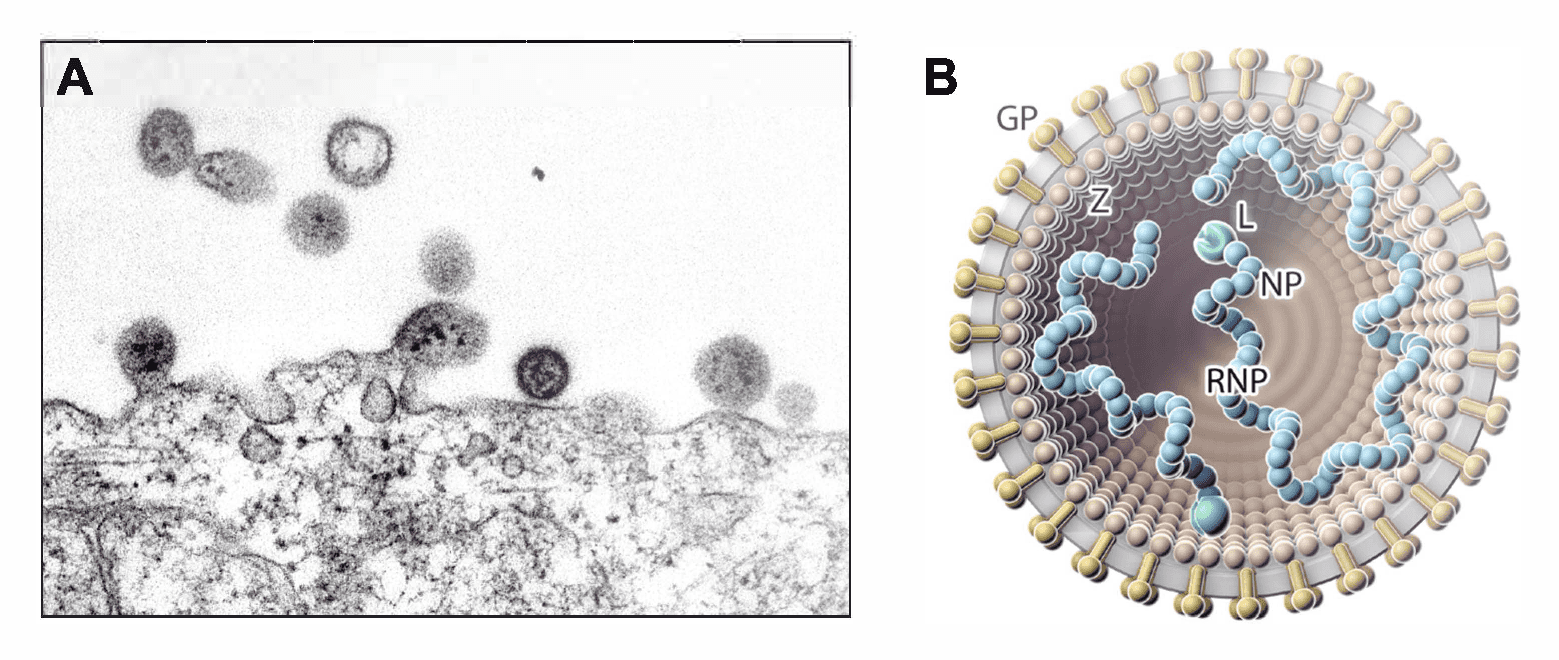 |
| Figure 1 Arenaviridae. (A) Electron micrograph of (mammalian) lymphocytic choriomeningitis virus particles, showing dark internal inclusion bodies (Latin: arena, sand) budding from an infected cell. (B) Illustration of a (mammalian) arenavirid particle showing the spherical and enveloped particle (grey) that is spiked with glycoproteins (GPs, gold) around a layer of zinc-binding proteins (Z, brown; missing in antennaviruses, hartmaniviruses, and innmoviruses). The small (S) and large (L; green) ribonucleoprotein (RNP) complexes inside the particle consist of nucleoprotein (NP; blue) and L protein. Antennavirus and innmovirus particles contain a third, medium (M), RNP complex. |
Physicochemical and physical properties
This information is mostly known for mammarenaviruses (see Mammarenavirus genus section).
Nucleic acid
Arenavirions typically contain two or three linear ambisense or negative-sense RNA segments that are encapsidated independently. These RNAs are uncapped (Leung et al., 1977) and contain a single non-templated G nucleotide at each of the 5′-ends (Garcin and Kolakofsky 1990, Raju et al., 1990, Shi et al., 2018). No poly(A) tracts are present at the 3′-ends. The termini of the RNAs have inverted complementary strands encoding transcription and replication initiation signals (Young and Howard 1983, Salvato et al., 1989, Harnish et al., 1993, Hepojoki et al., 2018).
Proteins
Arenavirids express three (hartmaniviruses, innmoviruses) or four (antennaviruses, mammarenaviruses, reptarenaviruses) structural proteins. The most abundant structural protein in virions is NP, which encapsidates the viral genomic segments. The least abundant protein is L protein, which contains an RdRP domain and mediates virus genome replication and transcription. Z, which is absent in antennaviruses, hartmaniviruses, and innmoviruses, is a matrix protein. Glycoprotein subunits GP1and GP2 are derived by post-translational cleavage of an intracellular GP precursor (GPC), the “glycoprotein-cell-associated” preprotein, by the cellular subtilisin kexin isozyme-1/site 1 protease (SKI-1/S1P). A third GPC cleavage product, SSP, stays attached to the GP complex in hartmaniviruses and mammarenaviruses but not in reptarenaviruses. The GP structure of antennaviruses and innmoviruses is unknown (Buchmeier et al., 1987, Lenz et al., 2001, Eichler et al., 2003, Kunz et al., 2003, York et al., 2004, Bederka et al., 2014, Koellhoffer et al., 2014, Hepojoki et al., 2018, Shi et al., 2018, Chen et al., 2021, Katz et al., 2022).
Lipids
This information is only known for mammarenavirions (see Mammarenavirus genus section).
Carbohydrates
This information is only known for mammarenavirions (see Mammarenavirus genus section).
Genome organization and replication
The arenavirid genome typically consists of two or three single-stranded RNA molecules named small (S), medium (M), and large (L). Some of these RNAs encode two proteins in non-overlapping ORFs of opposite polarities (ambisense coding arrangement) that are separated by non-coding intergenic regions (IGRs) (Figure 2 Arenaviridae). The S RNA encodes NP on the virus genome-complementary strand and, in many cases, GPC on the virus genome-sense strand. The L RNA encodes L protein on the virus genome-complementary strand and, in some cases, Z on the virus genome-sense strand. Antennaviruses, hartmaniviruses, and innmoviruses lack the Z ORF, and antennaviruses encode at least one protein of unknown function. The IGRs form one or more energetically stable stem–loop (hairpin) structures that function in structure-dependent transcription termination and in virion assembly and budding.
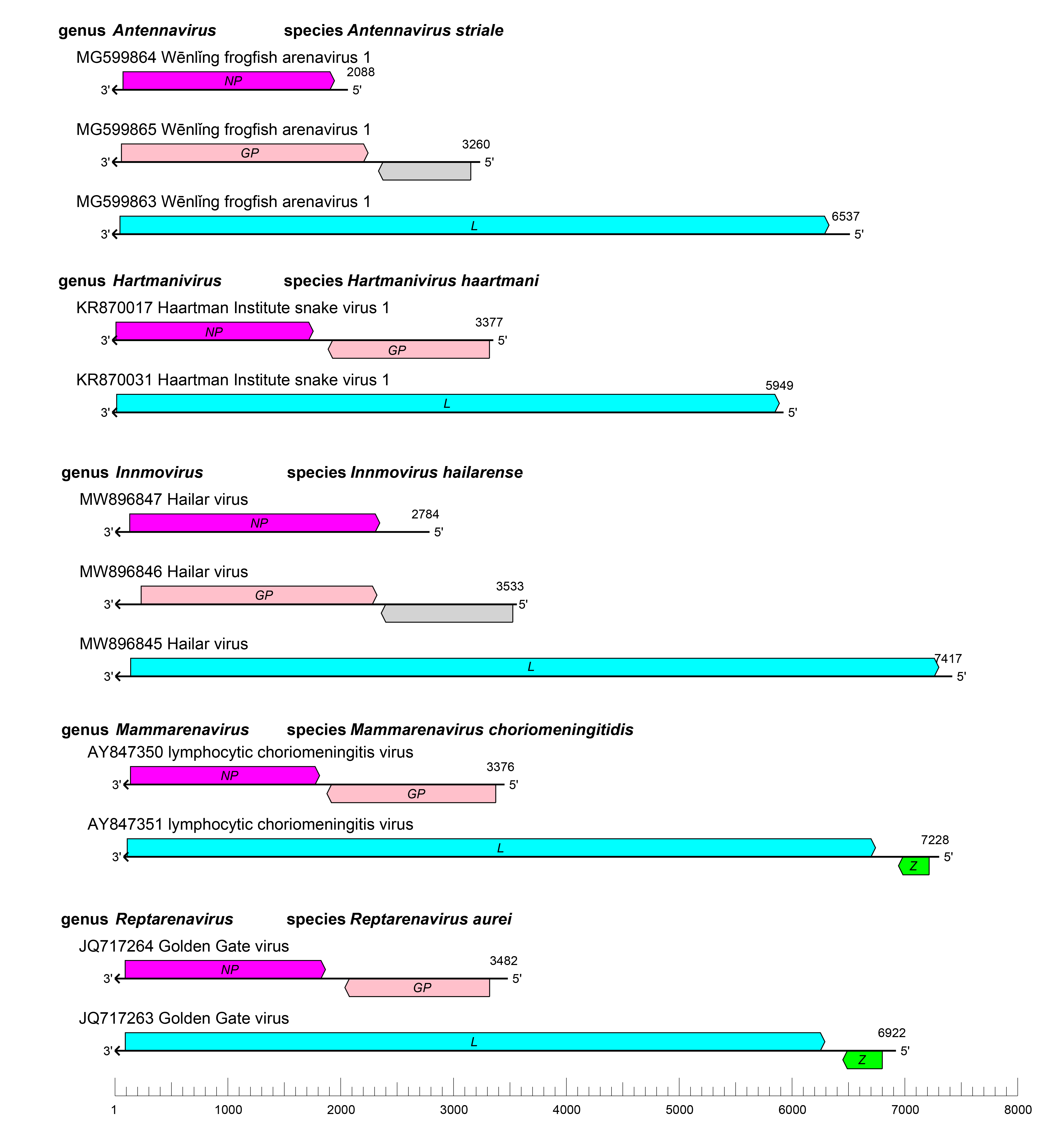 |
| Figure 2 Arenaviridae. Schematic representation of the bi-segmented or tri-segmented arenavirid genome organization (one example per genus). The 5′- and 3′-ends of all RNA segments are complementary at their termini, likely promoting the formation of circular ribonucleoprotein (RNP) complexes within the virion. GP, glycoprotein gene; L, large protein gene; NP, nucleoprotein gene; Z, zinc-binding protein gene. |
Arenavirid infection starts with attachment to cell-surface receptors or attachment factors, such as dystroglycan 1 in case of LASV and LCMV and transferrin receptor 1 in the case of several South American mammarenaviruses, followed by entry via the endosomal route (Glushakova and Lukashevich 1989, Borrow and Oldstone 1994, Cao et al., 1998, Martinez et al., 2007, Radoshitzky et al., 2007, Vela et al., 2007, Raaben et al., 2017) (Figure 3 Arenaviridae). pH-dependent fusion with late endosomes releases the virion RNP complex into the cytoplasm. For some mammarenaviruses, this fusion event involves a pH-dependent switch to an intracellular receptor, lysosomal associated membrane protein 1 (LAMP1) in the case of LASV and the CD164 molecule in the case of LCMV (Jae et al., 2014, Bakkers et al., 2022). The virus RNP directs both RNA genome replication and gene transcription (Meyer et al., 2002). During replication, L protein reads through the IGR transcription–termination signal and generates uncapped antigenomic and genomic RNAs (Leung et al., 1977). Because these RNAs contain single non-templated Gs at their 5′-ends (Garcin and Kolakofsky 1990, Raju et al., 1990), replication initiation might involve a slippage mechanism of L protein on the nascent RNA (Garcin and Kolakofsky 1992). In the case of ambisense coding arrangements, only mRNAs encoding NP or L protein can be synthesized from genomic RNAs and transcription of mRNAs encoding GPC or Z occurs only after the first round of virus replication, during which S and L antigenomes are produced.
Virus proteins are synthesized from subgenomic capped mRNAs that lack 3′-terminal poly(A) tracts (Singh et al., 1987, Southern et al., 1987, Meyer and Southern 1993). The 5′-ends of virus mRNAs contain several non-templated bases, suggesting that arenavirids use a cap-snatching mechanism similar to that used by other viruses of the negarnaviricot subphylum Polyploviricotina (Garcin and Kolakofsky 1990, Raju et al., 1990, Meyer and Southern 1993). Cap-snatching requires an endonuclease present in the N-terminal part of L, which cleaves cellular mRNAs to generate a cap leader that is subsequently used to prime arenavirid transcription. The 3′-ends of the mRNAs have been mapped to locations in the IGRs.
Virion budding, at least in case of mammarenaviruses, occurs from the cellular plasma membrane, thereby providing the virion envelope (Dalton et al., 1968, Perez et al., 2003, Strecker et al., 2003, Eichler et al., 2004).
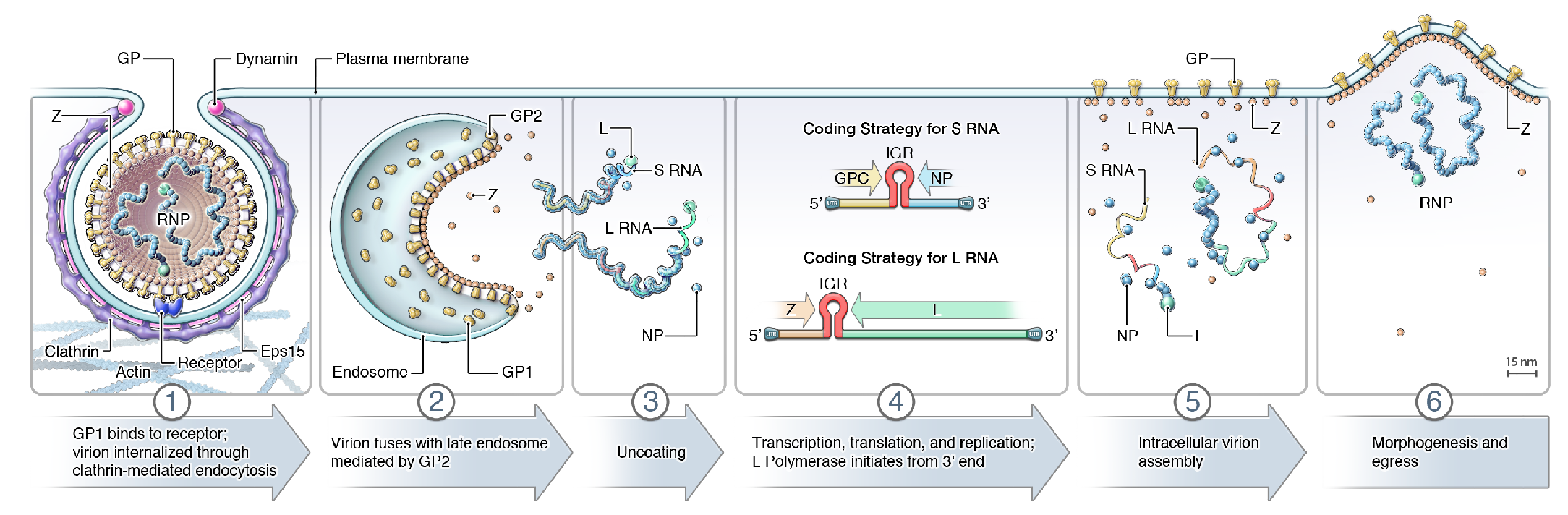 |
| Figure 3 Arenaviridae. Lifecycle of arenavirids. (1) Virion uptake; (2) virus-cell membrane fusion; (3) uncoating; (4) transcription, translation, and replication; (5) virion assembly; and (6) virion budding. GP, glycoprotein; IGR, intergenic region; L, large protein; NP, nucleoprotein; RNP, ribonucleoprotein; Z, zinc-binding protein. Note that antennaviruses, hartmaniviruses, and innmoviruses do not encode Z, possibly indicating that the lifecycle as depicted here is mammarenavirus-specific. |
Biology
Arenavirids are ecologically diverse: They have been detected in fish (antennaviruses) (Shi et al., 2018); bats, pikas, rodents, ticks, and a eulipotyphla (mammarenaviruses) (Downs et al., 1963, Sayler et al., 2014, Cui et al., 2023, Luo et al., 2023, Reuter et al., 2023); snakes (reptarenaviruses, hartmaniviruses) (Stenglein et al., 2012, Hetzel et al., 2013, Hepojoki et al., 2015, Hepojoki et al., 2018); and river sediment samples (innmoviruses). The geographic distribution of arenavirids overlaps with the distribution of their hosts. Most known mammalian arenavirids infect rodents of one or a few species and are therefore geographically constrained to their hosts, but LCMV, which infects the ubiquitous house mouse (Mus musculus Linnaeus, 1758), is distributed globally (Childs 1993). The natural distribution of reptilian arenavirids is unknown because they have only been detected in captive snakes (Stenglein et al., 2012, Hetzel et al., 2013, Hepojoki et al., 2015, Hepojoki et al., 2018) and recently in wild boid snakes in Costa Rica (Alfaro-Alarcón et al., 2022). A diverse range of vertebrate cell lines are permissive to mammarenavirus infection in vitro; some reptilian cell lines support replication of reptilian arenavirids (Lukashevich et al., 1983, Stenglein et al., 2012, Hepojoki et al., 2018) in line with a recent description of a possible arenavirid in a lizard (Harding et al., 2022). The ecology of innmoviruses remains to be determined.
Antigenicity
Systematic antigenicity studies have only been reported for mammarenavirions (see section on Mammarenavirus genus page).
Derivation of names
Arenaviridae: from the Latin arenosus meaning “sandy” and arena meaning “sand,” in recognition of the “sandy” appearance of mammarenavirus particles observed in electron-microscopic thin sections (Rowe et al., 1970a).
Antennavirus: from Antennarius striatus, the fish species to which the presumed host of Wēnlǐng frogfish arenavirus 1 (WlFAV1) and Wēnlǐng frogfish arenavirus 2 (WlFAV2), striated frogfish, has been assigned (Shi et al., 2018).
Hartmanivirus: from Haartman Institute, located at the University of Helsinki in Finland, the place where Haartman Institute snake virus 1 (HISV-1) was discovered (Hepojoki et al., 2015).
Innmovirus: from Inner Mongolia Autonomous Region (内蒙古自治区; Nèiménggǔ zìzhìqū), China, where Hailar virus (HLRV) was discovered (Chen et al., 2021).
Mammarenaviridae: from the Latin mamma, “udder or “breast”, referring to mammalian hosts of these viruses.
Reptarenavirus: from the Latin repere meaning “creep” or “crawl”, a reference to the reptilian hosts of reptarenaviruses (Radoshitzky et al., 2015).
Genus demarcation criteria
Classification of arenavirids is based on pairwise sequence comparison (PASC) of coding-complete genomes. Using the most current sequence dataset, S-segment and L-segment nucleotide sequence identities for viruses within the same genus need to be higher than 40 and 35%, respectively (Radoshitzky et al., 2015). Five genera have been established. Viruses assigned to a genus form a monophyletic clade in well-supported maximum-likelihood trees using complete L and NP nucleotide sequences and/or core L protein palm domain sequences. Use of L and NP for taxonomic purposes is justified by the presence of broadly conserved domains and the rarity of reassortment among genomic segments, at least in mammarenaviruses. Hence, coding-complete sequences of all genomic segments may be sufficient for arenavirus classification in the absence of a cultured isolate (Radoshitzky et al., 2015). However, classification also includes the consideration of phenotypic characters, such as significant differences in member virus genome architecture, virion antigenicity, and virus ecology (e.g., host range, pathobiology, and transmission patterns).
Relationships within the family
Phylogenetic relationships across the family have been established from maximum-likelihood trees generated using complete L protein amino-acid sequences (Figure 4 Arenaviridae). Phylogenetic relationships among viruses assigned to more closely related genera and within genera can also be established using other structural protein genes, including NP.
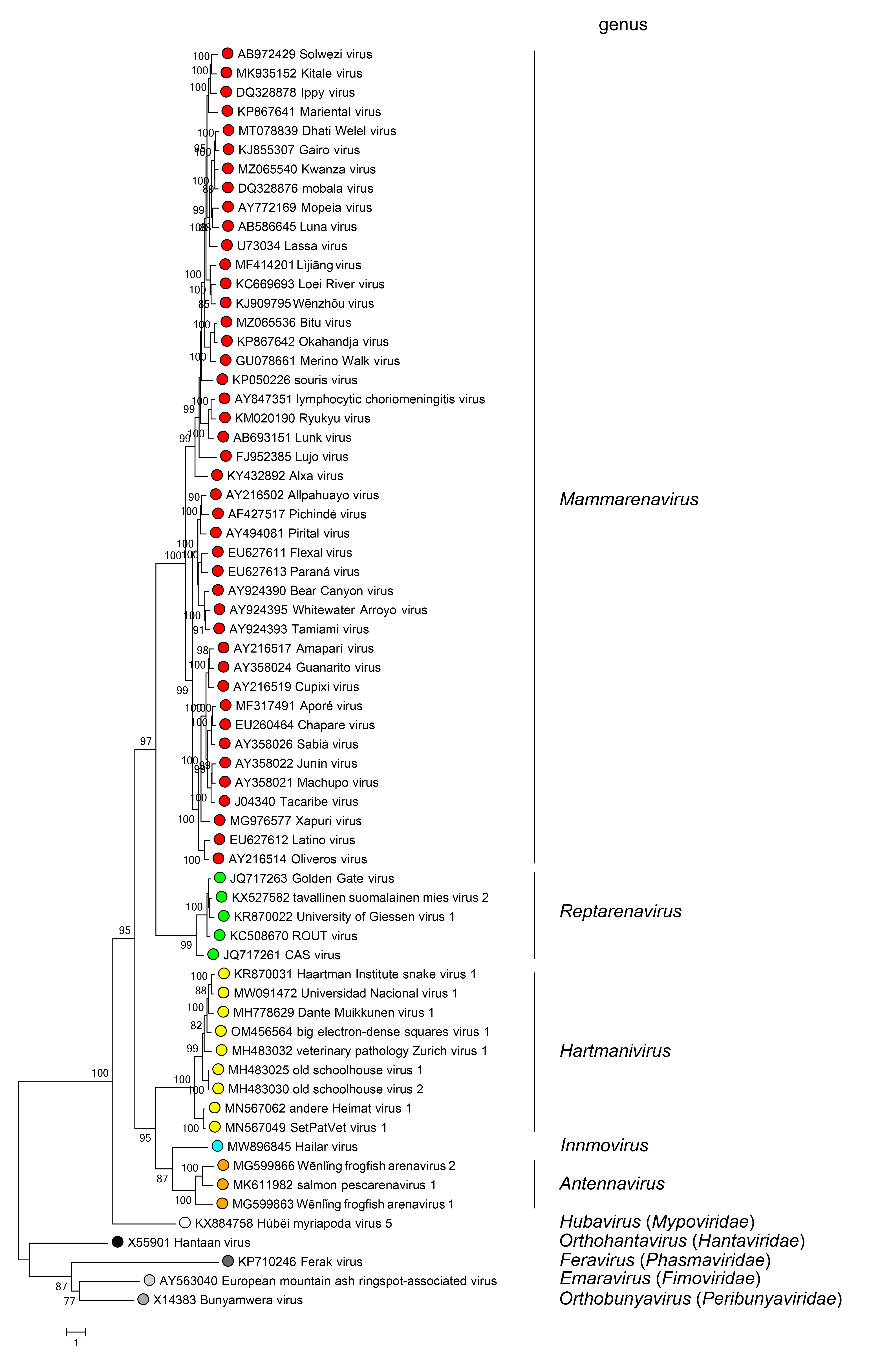 |
| Figure 4 Arenaviridae. Maximum-likelihood phylogenetic tree inferred from MAFFT alignment (Katoh and Standley 2013) of the complete L protein amino-acid sequences of 61 arenavirids assigned to the genera Antennavirus (cyan dots), Hartmanivirus (dark green dots), Innmovirus (yellow dot), Mammarenavirus (red dots), and Reptarenavirus (light green dots), along with representative viruses of other bunyaviral families (dots of other colors). The tree was generated by the IQ-TREE software v.1.6.12 (Trifinopoulos et al., 2016). IQ-TREE selects the best-fitting evolutionary model and then generates a maximum-likelihood tree. Branch supports were calculated using the ultrafast bootstrap method (1,000 bootstraps). The tree was visualized using FigTree (http://tree.bio.ed.ac.uk) and is mid-point rooted. |
Relationships with other taxa
Arenavirids are closely related to bunyaviral mypovirids, nairovirids, phenuivirids, and wupedevirids (Shi et al., 2016).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| anole arenavirus | S segment: BK061390*; L segment: BK061391* | (Harding et al., 2022) | |
| DF 20/00 virus | (Granzow et al., 2014) | ||
| DF 26/02 virus | (Granzow et al., 2014) | ||
| Hyriopsis cumingii Lea plague virus | HcPV | (Zhong et al., 2011, Carella et al., 2016) |
Virus names and abbreviations are not official ICTV designations.
*Coding region sequence incomplete.
Additional unclassified arenavirids that are probable members of existing genera are listed under individual genus descriptions.

