Subfamily: Orthoretrovirinae
Genus: Lentivirus
Distinguishing features
Virions have a distinctive morphology with a bar or cone-shaped core (nucleoid) (Pornillos and Ganser-Pornillos 2019). Viruses assemble at the cell membrane. The primate lentiviruses are distinguished from other viruses in the genus by the use of a chemokine receptor and the CD4 protein as receptors, the presence of a Nef protein, and the absence of a DU protein (Evans et al., 2013, Freed and Martin 2013).
Virion
Morphology
Virions have a distinctive, irregular icosahedral core that appears as a recognizable “cone-shape” morphology in electron micrographs.
Physicochemical and physical properties
See discussion under family description.
Nucleic acid
See discussion under family description.
Proteins
Approximate protein sizes are: matrix protein (MA) 17 kDa; capsid protein (CA) 24 kDa; nucleocapsid protein (NC) 7–11 kDa; p6 6 kDa; protease (PR) 14 kDa; reverse transcriptase (RT) 66 kDa; dUTPase (DU) (in all except the primate lentiviruses) 14 kDa; integrase (IN) 32 kDa; surface envelope protein (SU) 120 kDa; and transmembrane envelope protein TM 41 kDa. Detailed structural data are available for human immunodeficiency virus 1 (HIV-1) proteins MA, CA, NC, PR, RT, IN, SU and TM.
Genome organization and replication
The genome is about 9.3 kb (one monomer) (Figure 1. Lentivirus). In addition to the structural gag, pro, pol and env genes, there are additional genes, depending on the virus (e.g. for HIV-1: vif, vpr, vpu, tat, rev and nef) whose products are involved in regulation of synthesis and processing of virus RNA and combating host restriction factors. Most are located 3′ to gag-pro-pol and, at least in part, 5′ to env; one (nef in HIV-1) is 3′ to env. For other viruses there may be additional non-structural genes (e.g. vpx in human immunodeficiency virus 2, HIV-2). The tRNA primer is tRNALys-3. The long terminal repeat (LTR) is about 600 nt, of which the U3 region is 450 nt, R is 100 nt and the U5 region is 80 nt.
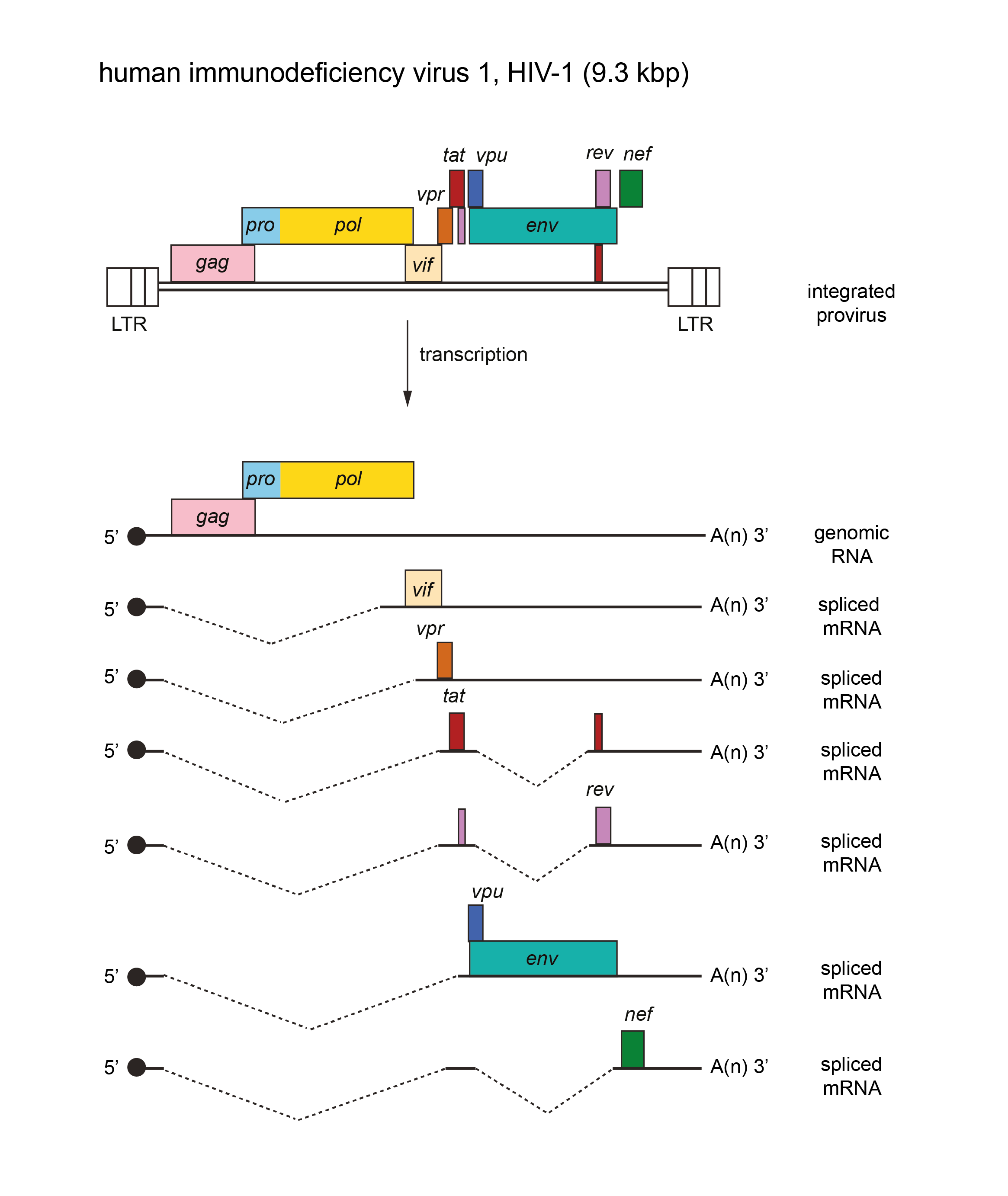 |
| Figure 1. Lentivirus. Lentivirus genome organization. The 9.3 kbp human immunodeficiency virus 1 (HIV-1) provirus is shown, with LTRs, protein-coding regions (gag, pro, pol, env, vif, vpr, vpu, tat, rev and nef) and transcripts (solid line arrows with protein names added) marked. The pro and pol genes are expressed as a result of a ribosomal frameshift. The coding regions in other members of the genus may occupy different reading frames. |
Biology
The viruses in the genus include exogenous viruses of humans and many other mammals that are transmitted horizontally and vertically (Evans et al., 2013, Freed and Martin 2013). Five groups of lentiviruses can be clustered on the basis of the hosts they infect (primates, sheep and goats, horses, cats and cattle) (Gifford 2012, Evans et al., 2013). Related endogenous retrovirus (ERV) sequences have been found in the genomes of lagomorphs, lemurs, mustelid carnivores and colugos (Gifford 2012). Lentiviruses are associated with a variety of diseases, including immunodeficiencies, neurological disorders and arthritis, whereas others appear non-pathogenic. No oncogene-containing member of this genus has been isolated.
Antigenicity
Some groups have cross-reactive Gag antigens (e.g. ovine, caprine and feline lentiviruses). Viruses related to isolates of Feline immunodeficiency virus have been isolated from other large felids (e.g. puma lentivirus), and antibodies to Gag antigens in lions and other large felids indicate the existence of additional viruses related to feline immunodeficiency virus (FIV) and ovine/caprine lentiviruses. There is limited cross-reactivity among primate lentiviruses in ELISA tests based on Gag components, but essentially none in those based on env gene products.
Species demarcation criteria
Species demarcation criteria have historically included differences in antigenic properties, differences in natural host range, and differences in pathogenicity, although these criteria have largely given way to the use of genome sequence divergence and the presence/absence of accessory genes. Lentiviruses of the same or closely related hosts typically form monophyletic clades within the genus Lentivirus. While the clades do not have a formal taxonomic designation, species demarcation within clades is based primarily on sequence divergence and the presence of any unique, derived features. For example, within the primate lentivirus clade, HIV-1 is distinguished from human immunodeficiency virus 2 (HIV-2) primarily on the basis of sequence divergence that exceeds 50% and the presence of the vpx gene in HIV-2. This difference reflects the different primate hosts that were sources for HIV-1 and HIV-2 (chimpanzees and sooty mangabeys, respectively) (Sharp and Hahn 2011).

