Subfamily: Orthoretrovirinae
Genus: Alpharetrovirus
Distinguishing features
Virus particles assemble at the plasma membrane and exhibit a “C-type” morphology. Viruses in the genus have an envelope TM protein with an internal fusion peptide flanked by two cysteines, whereas viruses in the other genera typically have an N-terminal fusion peptide. Many oncogene-containing members of the genus have been isolated.
Virion
Morphology
Virus particles assemble at the plasma membrane and exhibit a “C-type” morphology.
Physicochemical and physical properties
See discussion under family description.
Nucleic acid
See discussion under family description.
Proteins
Approximate protein sizes are: matrix (MA) 19 kDa ; p10 10 kDa ; capsid (CA) 27kDa; nucleocapsid (NC) 12 kDa; protease (PR) 15 kDa; reverse transcriptase (RT) 68 kDa; integrase (IN) 32 kDa; surface envelope protein (SU) 85 kDa; and transmembrane envelope protein (TM) 37 kDa. The SU and TM subunits of Env are covalently linked by an inter-subunit disulfide bond.
Genome organization and replication
The genome is about 7.2 kb (one monomer) (Figure 1. Alpharetrovirus). There are no known viral replication genes additional to gag, pro, pol and env. Some members of the genus also encode oncogenes, although these are usually in the context of defective genomes (Martin 2004). For example, the replication-defective alpharetroviruses can be distinguished from Rous sarcoma virus by the variable deletion of portions of the gag, pol and env genes and the presence of a unique oncogene in each species. The tRNA primer is tRNATrp. The long terminal repeat (LTR) is about 350 nt of which the U3 region is 250 nt, the R sequence is 20 nt and the U5 region is 80 nt.
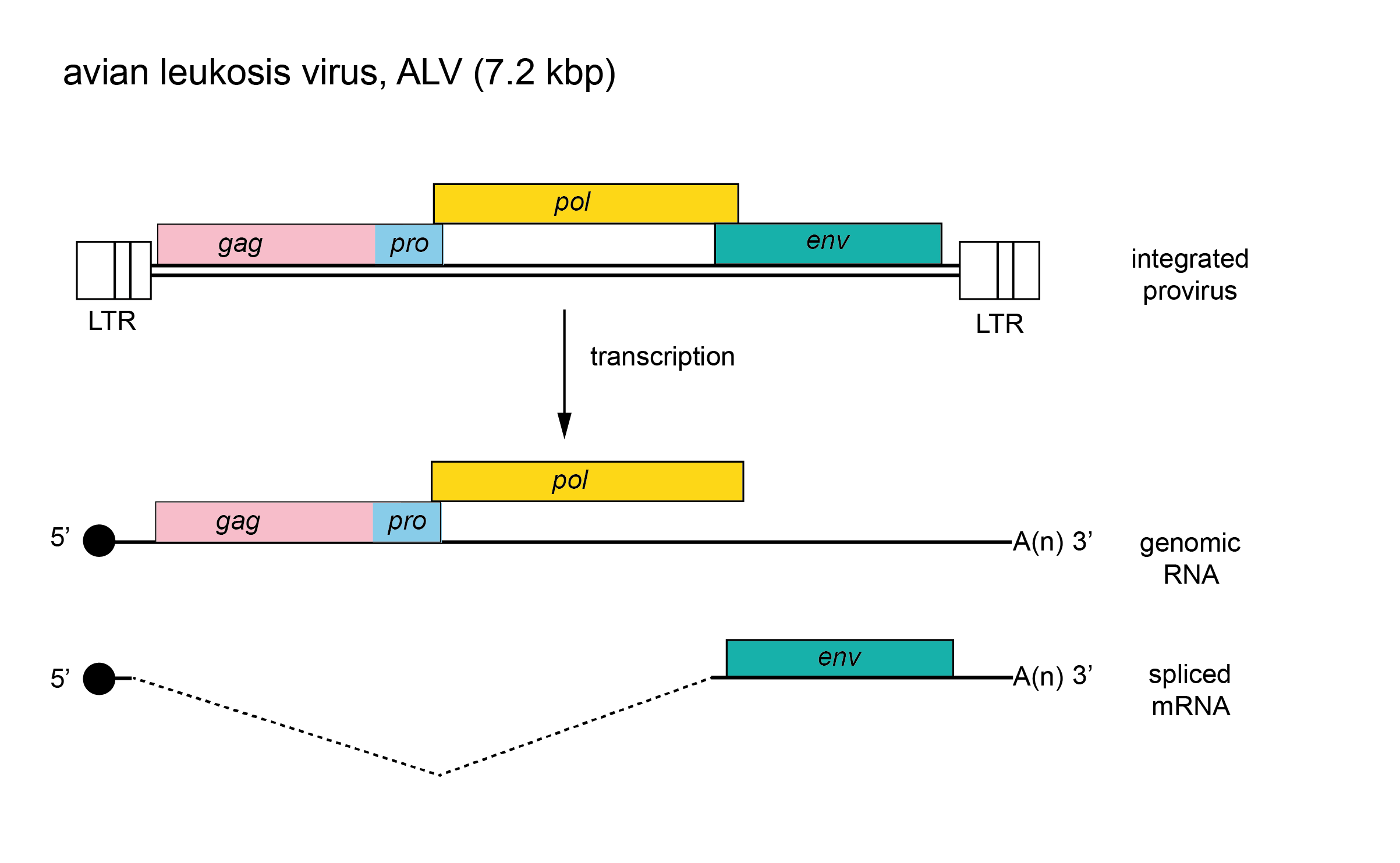 |
| Figure 1. . Alpharetrovirus. Alpharetrovirus genome organization. The 7.2 kbp avian leukosis virus (ALV) provirus is shown, with LTRs, genes (gag, pro, pol and env) and transcripts (solid line arrows with protein names added) marked. The pol gene is expressed as a result of a ribosomal frameshift. |
Biology
The viruses have a widespread distribution and include both exogenous (vertical and horizontal transmission) and endogenous viruses of chickens and some other birds. Isolates of avian leukosis virus (ALV) are classified into subgroups (e.g. -A, -J) by their distinct receptor usage (Barnard et al., 2006, Chai and Bates 2006, Lindemann et al., 2013). Related endogenous sequences are found in birds and mammals. Virus infections are associated with malignancies and some other diseases such as wasting, and osteopetrosis. Multiple alpharetroviral oncogenes have been described; for example, Rous sarcoma virus strains encode the src oncogene, whereas members of species Avian myeloblastosis virus encode the myb oncogene.
Species demarcation criteria
Species demarcation criteria include differences in genome sequences and differences in gene product sequences. For example, members of the species Avian leukosis virus can be readily distinguished from those of the species Rous sarcoma virus because they lack oncogene sequences while still encoding gag, pol and env. Host range, defined by SU interaction with a specific receptor, is more commonly used in defining strains within a species.

