Subfamily: Orthoretrovirinae
Genus: Gammaretrovirus
Distinguishing features
Viruses in the genus have simple genomes comprising just the canonical gag, pro, pol and env genes, with little or no evidence for additional regulatory genes. Instead of frameshifting, translation of the Gag-Pro-Pol polyprotein relies on readthrough of the Gag termination codon. In contrast to viruses in the Betaretrovirus and Lentivirus genera, the SU and TM subunits of Env are covalently linked by a disulfide bond that undergoes isomerization as part of the fusion and entry mechanism.
The mammalian gammaretroviruses include replication-competent viruses, which lack cell-derived oncogenes, and replication-defective viruses, which have acquired a variety of oncogenes from their hosts. Murine and feline transforming gammaretroviruses, which are invariably replication-defective, can be distinguished from members of the species Murine leukemia virus and from one another by the presence of a large variety of distinct cell-derived oncogenes (e.g. mos in isolates of Moloney murine sarcoma virus and sis in isolates of Woolly monkey sarcoma virus) and the characteristic loss of portions of gag, pol or env.
Collectively, gammaretroviruses have a very broad host range, and have been detected in, or isolated from, a wide variety of vertebrate hosts. Receptor use is correspondingly diverse, but with a very strong preference for proteins in the solute carrier (SLC) superfamily. As with alpharetroviruses, even closely related viruses may use distinct host cell receptors for viral entry (Greenwood et al., 2018, Overbaugh et al., 2001).
Virion
Virions exhibit a “C-type” morphology with barely visible surface spikes. They have a centrally located, condensed core. Capsid assembly occurs at the inner surface of the membrane at the same time as budding.
Physicochemical and physical properties
See discussion under family description.
Nucleic acid
See discussion under family description.
Proteins
Approximate protein sizes are: matrix protein (MA) 15 kDa; p12 12 kDa; capsid protein (CA) 30 kDa; nucleocapsid protein (NC) 10 kDa; protease (PR) 14 kDa; reverse transcriptase (RT) 80 kDa; integrase (IN) 46 kDa; surface envelope protein (SU) 70 kDa; and transmembrane envelope protein (TM) 15 kDa.
Genome organization and replication
The genome is about 8.3 kb (one monomer) (Figure 1. Gammaretrovirus). The pro-pol region is translated following ribosomal readthrough at the gag gene termination codon. Some gammaretroviruses additionally translate the gag region from an upstream initiation codon, yielding a larger, glycosylated form of Gag protein, glyco-Gag. There are no known additional genes. The tRNA primer is usually tRNAPro , although examples of tRNAGlu, tRNAGln and tRNAThr have been found among endogenous mouse viruses. The long terminal repeat (LTR) is about 600 nt, of which the U3 region is 500 nt, the R sequence is 60 nt and the U5 region is 75 nt.
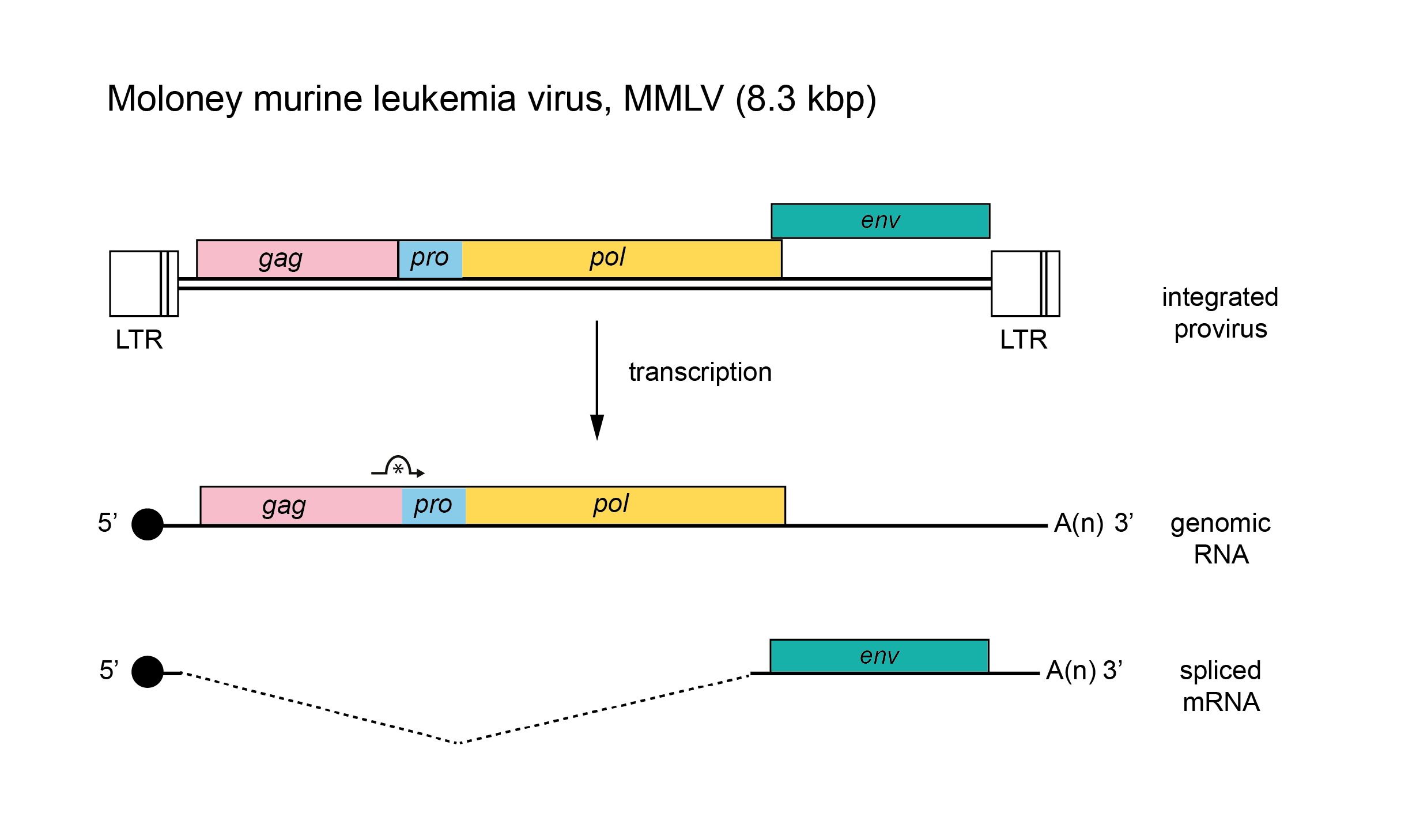 |
| Figure 1. Gammaretrovirus. Gammaretrovirus genome organization. The 8.3 kb provirus of Moloney murine leukemia virus (MMLV) is shown, with LTRs, protein-coding regions (gag, pro, pol and env) and transcripts (solid line arrows with protein names added) marked. The asterisk under a curved arrow indicates a ribosomal readthrough site. |
Biology
The viruses are associated with a variety of diseases, including malignancies, immunosuppression, and neurological disorders, and many oncogene-containing members of the mammalian and reticuloendotheliosis virus groups have been isolated (Rosenberg and Jolicoeur 1997). Viruses resulting from recombination between exogenous and endogenous viruses are frequently encountered. Gammaretroviruses are widely distributed with many mammals having both exogenous viruses transmitted vertically and horizontally, as well as endogenous viruses. For example, koala retrovirus (KoRV) is found in both exogenous and endogenous forms and appears to be actively endogenizing in Australian koala populations. The reticuloendotheliosis viruses (REV) comprise a few isolates from birds, but share features with several mammalian retroviruses suggesting a possible inter-class transmission. Interestingly, REV proviruses have been found in field samples integrated into the genomes of avian DNA viruses, including Marek’s disease virus (Gallid alphaherpesvirus 2, Herpesviridae) and fowlpox virus (Avipoxvirus, Poxviridae).
Species demarcation criteria
Species demarcation criteria include differences in genome sequence and presence of viral oncogenes, differences in antigenic properties, differences in natural host range and differences in pathogenicity. Members of the species Murine leukemia virus can be distinguished from isolates of Gibbon ape leukemia virus, for example, by sequence divergence, and only show limited antigenic cross-reactivity in ELISA assays.

