Book: Papillomaviridae
Subfamily: Firstpapillomavirinae
Genus: Chipapillomavirus
Distinguishing features
Members of this genus are associated with skin plaques in carnivore hosts.
Virion
See discussion under family description.
Genome organization and replication
See discussion under family description.
Biology
See discussion under family description.
Species demarcation criteria
Putative novel papillomavirus genome with complete genome sequence data available and that is <70% related to papillomaviruses within the genus (Figure 1. Chipapillomavirus).
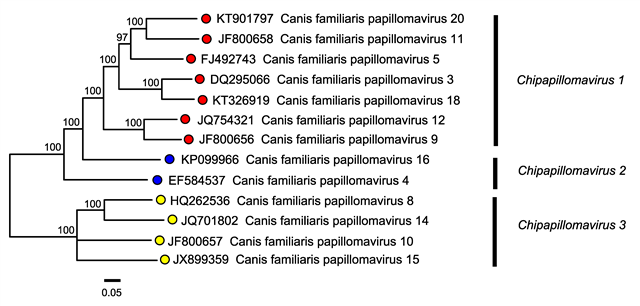 |
| Figure 1. . Chipapillomavirus. Phylogenetic tree of members of the genus Chipapillomavirus. The E1, E2, L2, and L1 nucleotide sequences of 343 papillomavirus isolates including representatives of all species and genera within the Papillomaviridae family were aligned as amino acid sequences using MUSCLE v7.221 (Edgar 2004). JModeltest2 (Darriba et al., 2012) was used to determine the optimal model of evolution (GTR + I + G) for the concatenated nucleotide sequences. Maximum likelihood (ML) trees were constructed using RAxML MPI v8.2.9 (Stamatakis 2006) implementing the GTR substitution model. ML bootstrap analysis used the autoMRE-based stopping criterion in RAxML. Following tree construction (tree available in the Resources section of the Papillomaviridae Report), the subtree corresponding to the genus Chipapillomavirus was isolated. Tips are labelled with virus names and accession numbers; nodes are labelled with bootstrap support values. |

