Family: Flaviviridae
Genus: Hepacivirus
Summary
The best characterised member of the Hepacivirus genus is hepatitis C virus (HCV), classified as a member of the species Hepacivirus hominis (Smith et al., 2016). HCV infects humans and is an important aetiological agent of chronic hepatitis. A further hepacivirus, GB virus-B (GBV-B), highly divergent in sequence from HCV, was first described in tamarins in 1995 in which it establishes acute infections and liver pathology comparable to that of HCV (Simons et al., 1995b). Several other hepaciviruses have recently been described that infect Old World colobus monkeys, various rodents and bats species, European and African cattle, horses and donkeys (Sibley et al., 2014, Walter et al., 2017, Corman et al., 2015, Baechlein et al., 2015, Kapoor et al., 2011, Burbelo et al., 2012, Kapoor et al., 2013b, Drexler et al., 2013, Firth et al., 2014, Quan et al., 2013). These viruses have been assigned to further hepacivirus species and show distinct host ranges - Hepacivirus equi (non-primate hepacivirus, NPHV) infecting horses and possibly dogs, Hepacivirus platyrrhini (GBV-B) infecting tamarins and potentially other New World primates, Hepacivirus colobi infecting colobus monkeys,Hepacivirus peromysci, Hepacivirus myodae, Hepacivirus ratti, Hepacivirus norvegici, Hepacivirus rhabdomysis and Hepacivirus glareoli infecting various species of rodents, Hepacivirus macronycteridis, Hepacivirus vittatae and Hepacivirus otomopis infecting bats and Hepacivirus bovis infecting cows. A currently unclassified, more divergent hepacivirus-like sequence has been assembled from liver tissue of a gracile shark (Wenlin shark virus).
Collectively, these other hepaciviruses are much less well characterised virologically and clinically than HCV (Hepacivirus hominis) and the descriptions below primarily refer to HCV; any available information on other members is referred to where appropriate.
A Table with current (MSL38) and previous (MSL37) species names is available at Species names: Flaviviridae.
Distinguishing features
Hepatitis C virus (HCV) is transmitted between humans, principally via exposure to contaminated blood; the transmission routes of other hepaciviruses are poorly understood but are unlikely to be parenteral. There is no known invertebrate vector for HCV or any other hepacivirus. Hepaciviruses differ from members of the genera Orthoflavivirus and Pestivirus by their limited ability to be propagated in cell culture; only a few adapted strains of HCV, including JFH1, efficiently infect the only susceptible cultured cell line, human hepatoma cell line (Huh7). Cell culture of other hepaciviruses has not been achieved to date. In the HCV precursor protein, the NS2-3 junction is auto-catalytically cleaved by Zn-dependent NS2-3 protease activity; a similar mechanism is likely for other hepaciviruses.
Virion
Morphology
Virions of HCV are about 50 nm in diameter, as determined by filtration and electron microscopy. They are spherical in shape with a lipid envelope, as determined by electron microscopy and inactivation by chloroform. The viral core is spherical and about 30 nm in diameter. Detailed structural properties of HCV or other hepaciviruses have not been determined.
Physicochemical and physical properties
Virion Mr has not been determined. The buoyant density in sucrose of HCV is predominantly about 1.06 g cm−3 for virus recovered from serum during acute infections while more dense forms (ca. 1.15–1.18 g cm−3) predominate when recovered from the serum of chronically infected individuals (Thomssen et al., 1993). Lower density banding results from physical association of the virion with serum very-low-density lipoproteins (VLDLs)(Thomssen et al., 1992). Higher density virions are those bound to serum antibodies. Of these different particle types found in humans, the lowest density particles are the most infectious (paricles < 1.1 g cm-3)(Bradley et al., 1991). A buoyant density range in isosmotic iodixanol gradients of 1.01–1.10 g cm−3 has been measured for HCV recovered from hepatoma cells infected with HCV. The S20,w is equal to or greater than 150S. The virus is stable in buffer at pH 8.0–8.7. Virions are sensitive to heat, organic solvents and detergents (Feinstone et al., 1983).
Nucleic acid
Hepacivirus virions contain a single positive-sense, infectious ssRNA (Figure 1. Hepacivirus). The genome of HCV is about 9.6 kb, while for other hepaciviruses the range is 8.9–10.5 kb. The 5′-NCR of HCV possesses a type IV IRES (Honda et al., 1999) of approximately 340 nt. The divergent GBV-B, a member of Hepacivirus platyrrhini, has a genome organization similar to HCV, but with a more extensive 5′-NCR (445 nt) also containing a type IV IRES (Muerhoff et al., 1995). Type IV IRESs are similarly present in members of most other hepacivirus species although the rodent hepaciviruses that are members of Hepacivirus myodae and Hepacivirus glareoli possess an IRES showing sequence homology to those of pegiviruses (Smith et al., 2016, Drexler et al., 2013). The 3′-NCR of HCV contains a sequence-variable region of about 50 nt, a polypyrimidine-rich region (average of 100 nt), and a highly conserved 98 nt 3′-terminal region with three stem-loop RNA secondary structures (Tanaka et al., 1995) The 3′-NCRs of members of other hepacivirus species show little or no conservation in sequence or predicted RNA structure to that of HCV. There are at least two seed sites in the HCV 5′-NCR for the liver abundant microRNA miR-122; this virus-host interaction is required for efficient HCV replication (Jopling et al., 2005). One or two miR-122 seed sites are present in hepaciviruses of other species (Smith et al., 2016).
Proteins
The HCV virion comprises at least three proteins: the nucleocapsid core protein C (p19-21), and two envelope glycoproteins, E1 (gp31) and E2 (gp70). An additional protein, p7 (believed to have properties of an ion channel protein important in viral assembly) is incompletely cleaved from a precursor of E2 to yield E2-p7 and p7 (Shanmugam and Yi 2013) but it is not known whether these are virion structural components. In GB virus-B, a corresponding protein, p13, is cleaved to p7 and p6 proteins (Takikawa et al., 2006). The two envelope glycoproteins can associate as non-covalent heterodimers; recent data, however, indicate that they are covalently linked in virions (Vieyres et al., 2010). Nonstructural proteins include NS2, a 21 kDa protein that, before cleavage, is part of a Zn-dependent cysteine protease that bridges NS2 and NS3 and mediates autocatalytic cleavage of the NS2/NS3 junction, and is involved with virus assembly and release, NS3, a 70 kDa protein with additional serine protease, helicase and NTPase activities; the NS3 protease cleaves the remaining junctions between nonstructural proteins, NS4A , a 6 kDa cofactor essential for trans NS3 serine protease activity, NS4B, a 27 kDa protein that induces a membranous replication complex at the endoplasmic reticulum, NS5A, a serine phosphoprotein of unknown specific function, but critical for viral replication and assembly, that exists in 56 and 58 kDa forms, depending on the degree of phosphorylation, and NS5B, a 68 kDa protein with RdRP activity.
The genomes of other hepacivirus species show similar organisations to those of HCV and GB virus-B, with predicted cleavage sites in the coding region potentially producing core, E1, E2, p7/p13, NS2, NS3, NS4A, NS4B, NS5A and NS5B proteins homologous to and comparable in size to those of HCV and GB virus-B. One exception is a large insertion of intrinsically disordered amino acid sequence in the NS5A gene of the colobus monkey hepacivirus (Hepacivirus colobi) (Lauck et al., 2013).
Lipids
Hepacivirus virions have a lipid bi-layer envelope. Historically, based on the removal of the viral envelope and loss of infectivity of HCV following exposure to solvents or detergents (Feinstone et al., 1983), the presence of lipids was inferred. Recently, it has become apparent that the host lipid metabolism plays a critical role in the virus life cycle of HCV and likely other hepaciviruses.
Carbohydrates
The E1 and E2 glycoproteins of all hepaciviruses contain numerous N-linked glycosylation sites (1–4 in E1, 2–11 in E2), and carbohydrate is associated with the products of these two genes. E1 and E2 are transmembrane, type I glycoproteins, with C-terminal retention signals that anchor them within the lumen of the endoplasmic reticulum. These signals are apparently masked when budding occurs allowing the virion to move through the secretory pathway. Recent data obtained in culture systems indicate that N-linked glycans of HCV E1 remain in the high-mannose chains lacking complex carbohydrate, whereas those of E2 are modified (Op De Beeck et al., 2004). Glycosylation influences E1–E2 heterodimer formation, folding and assembly and the release of virions (Meunier et al., 1999).
Genome organization and replication
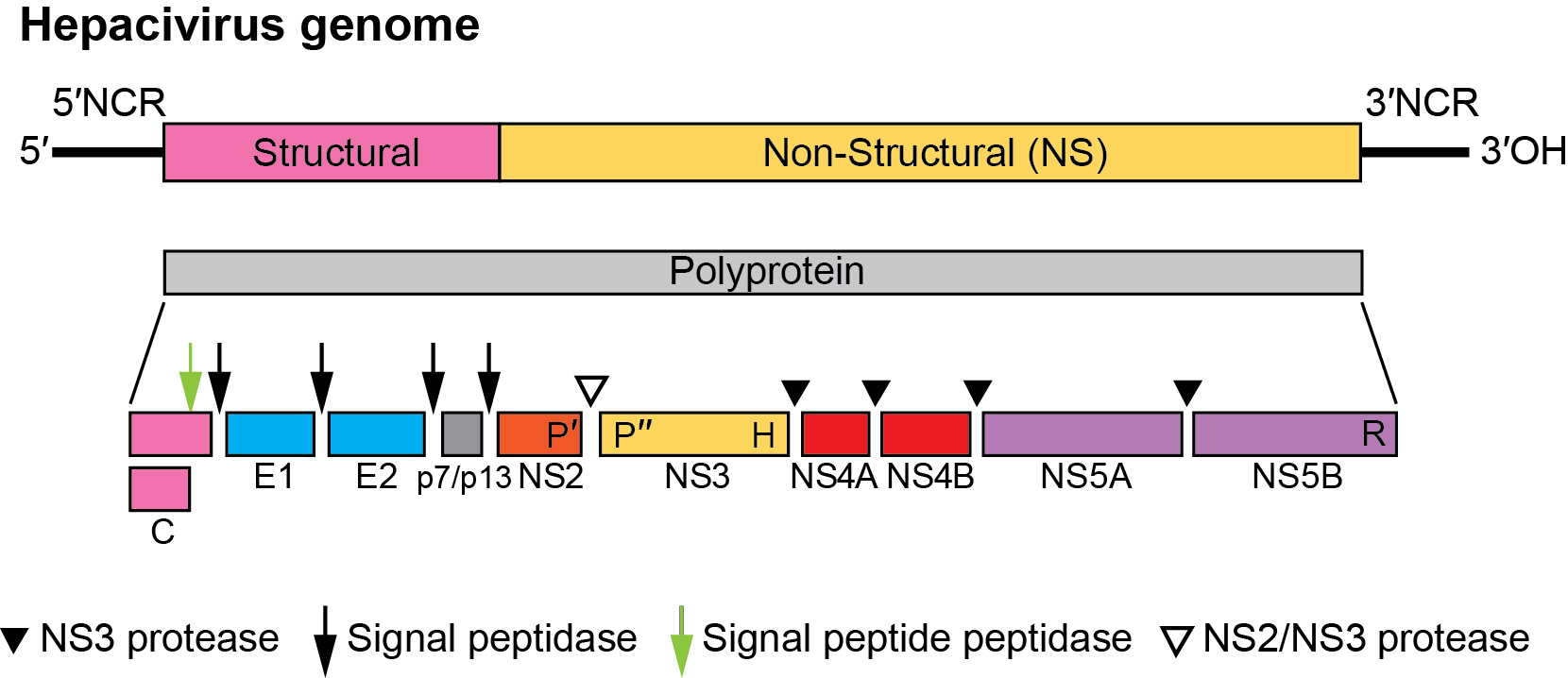
The hepacivirus genome contains a single large ORF encoding a polyprotein of about 3,000 aa (Figure 1. Hepacivirus). The gene order is 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′. Immediately downstream of the three structural proteins (C, E1, E2) is a small protein, p7 in HCV, p13 in GB virus-B and comparably small predicted proteins in other hepaciviruses, followed by the nonstructural proteins in the 3′-portion of the ORF. Replication occurs in association with intracytoplasmic membranes. Replicative forms of HCV and of the recently described non-primate hepacivirus (NPHV, Hepacivirus equi; (Pfaender et al., 2015)) have been detected in liver tissue. The genomic RNA is translated into a polyprotein that is rapidly processed both co- and post-translationally by host and viral proteases. Translation initiation occurs via an IRES within the 5′-NCR. Translocation of the structural glycoproteins to the endoplasmic reticulum probably occurs via internal signal sequences. Cleavage of the structural proteins is mediated by host cell signal peptidases, and signal peptide peptidase. With the exception of the p7/NS2 signalase cleavage, viral proteases cleave all non-structural protein junctions. Virus assembly is believed to occur by budding into vesicles from the endoplasmic reticulum.
|
|
|
Figure 1. Hepacivirus. Hepacivirus genome organization (not to scale) and polyprotein processing. For members of the species Hepacivirus hominis, the RNA is about 9.6 kb. The 5′-NCR is about 340 nt, the 3′-NCR about 250 nt, and the ORF about 9 kb. HCV has a p7 protein between E2 and NS2. The host and viral proteases involved in cleavage of the polyprotein are indicated. The cleavage by host signal peptide peptidase (at the C-terminus of the core protein) is indicated by a green arrow; the cleavages by host signal peptidase (remaining sites) are indicated by filled arrows. The locations of the NS2-3 protease, NS3 protease, NS3 RNA helicase and NS5B RdRP are indicated by P′, P″, H and R, respectively. |
Biology
Host range
Humans are the natural host and apparent reservoir of HCV although the virus can be transmitted experimentally to chimpanzees. No other natural host has been identified. The natural host for GB virus-B is not known although it can experimentally infect the New World primate species marmosets and tamarins and the more distantly related owl monkeys (Bukh et al., 2001). Members of other species of hepaciviruses infect a wide range of other mammalian species, both wild and domestic, including colobus monkeys (Sibley et al., 2014), cows (Baechlein et al., 2015), horses (Burbelo et al., 2012), donkeys (Walter et al., 2017) and a range of rodent and bat species (Kapoor et al., 2011, Kapoor et al., 2013b, Drexler et al., 2013, Firth et al., 2014, Quan et al., 2013). The host specificity of these variants for their mammalian hosts is undetermined, although at least one rodent species, the bank vole (Moyodes glareolus) can be infected with two highly divergent hepacivirus variants (RMU10-3382/GER/2010 and NLR07-oct70/NEL/2007)(Drexler et al., 2013) belonging respectively to the species Hepacivirus J and Hepacivirus F. This observation is indicative of a potential degree of cross-species transmission.
Transmission
HCV is transmitted almost exclusively by parenteral exposure to blood, blood products and objects contaminated with blood. Effective screening of blood donors and implementation of inactivation procedures have virtually eliminated the transmission of HCV by blood and blood products, but other routes of exposure, principally by blood-contaminated syringes, are now the most important recognized risk factors. Sexual and mother-to-child transmission has been documented but is relatively uncommon. Other routes of transmission are suspected for members of other hepacivirus species; in a recent study of thoroughbred horses, evidence for vertical transmission of NPHV was obtained from one of four mares to their foals, while further infections occurred in the post-natal period (Gather et al., 2016).
Geographical distribution
HCV has a worldwide distribution with about 3% of the world population infected with HCV, equivalent to 170 million chronic infections, with 3–4 million new infections each year. Antibody prevalences are 0.1–2% in developed countries but as high as 20% in some developing countries, possibly reflecting the historic use of contaminated needles and syringes. Horses infected with members of Hepacivirus equi have been reported from four continents with viraemia frequencies ranging from 3–10%, indicative of a wide geographical distribution (Burbelo et al., 2012, Pfaender et al., 2015, Lyons et al., 2012, Figueiredo et al., 2015, Lu et al., 2016, Matsuu et al., 2015).
Pathogenicity
The effects of HCV infection in humans range from subclinical to acute and chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. Persistent infection occurs in 60–80% of cases, and in about 20% of the cases the infection progresses over many years to chronic active hepatitis and cirrhosis. Patients with liver cirrhosis have an approximately 5% risk per year of developing hepatocellular carcinoma.
Persistent HCV infection has been linked by epidemiological studies to primary liver cancer, cryptogenic cirrhosis and some forms of autoimmune hepatitis. Extrahepatic manifestations of HCV infection include mixed cryoglobulinemia with associated membrano-proliferative glomerulonephritis and, possibly, porphyria cutanea tarda, Sjögren’s-like syndromes and other autoimmune conditions.
Similarly to HCV infections in humans, GB virus-B causes hepatitis and replicates in the liver of tamarins and owl monkeys, but infection is self-limited and has not been demonstrated in humans or chimpanzees. Only one strain of GB virus-B has been identified to date, in contrast to thousands of often quite divergent variants of HCV. The pathogenicity of other hepaciviruses infecting non-human primates, rodents, bats, cattle, and horses is poorly characterized although the presence of miR-122 seed sites in each species characterised to date predicts widespread hepatotropism among members of this genus (Jopling et al., 2005). Recent evidence suggests that infection of horses with NPHV may be associated with higher rates of spontaneous clearance than observed for HCV in humans (Pfaender et al., 2015, Lyons et al., 2014) and only mild inflammatory liver disease (Pfaender et al., 2015) with minimal elevation of liver enzymes (Lyons et al., 2014). Infection outcomes have been suggested to be dependent on the breed and age of the horse (Ramsay et al., 2015).
Cell tropism
HCV has been reported to replicate in several cell lines derived from hepatocytes and lymphocytes, but virus growth has only been sufficient for practical application of these systems in a human hepatoma cell line, Huh7 cells and derivatives thereof. In vivo, HCV replicates in hepatocytes and possibly lymphocytes. The cellular or tissue tropism of other hepaciviruses is poorly characterized although there is evidence that GB virus-B, NPHV, and bovine hepacivirus are hepatotropic; the presence of binding sites for miR-122 (Jopling et al., 2005) in other described hepaciviruses is consistent with widespread hepatotropism, given the restriction of expression of this miRNA to liver tissue.
Antigenicity
Virus-specific antibodies to recombinant-expressed structural proteins of HCV (C, E1 and E2) and nonstructural proteins (principally NS3, NS4 and NS5) have been detected in individuals infected with HCV. Both linear and conformational epitopes are believed to be involved in the humoral immune response of the host to infection. Significant antigenic diversity throughout the genome is reflected in heterogeneity in the humoral immune response. In HCV, high variability is found in the N-terminal 27 aa of E2 (hypervariable region 1; HVR1). The HVR1 contains an HCV neutralization epitope and escape variants of HVR1 are positively selected by the host humoral immune response (Mondelli et al., 2003). Other neutralization epitopes have been identified in E2 outside of HVR1 in E2 and also in E1 (Fafi-Kremer et al., 2012, Edwards et al., 2012, Keck et al., 2004). With the development of intra- and intergenotypic genotype 1-7 JFH1-based recombinant viruses with strain-specific structural proteins, it is now possible to carry out in vitro virus neutralization assays to address the antigenic diversity of HCV (Gottwein et al., 2011). Serological responses to GB virus-B infection are less well-characterized; antibodies to NS3 become detectable after a delay of several weeks in acutely infected tamarins but titres decline rapidly following virus clearance (Beames et al., 2001, Schaluder et al., 1995).
Cell-mediated immune responses to all HCV proteins have been detected (Klenerman and Thimme 2012); it is believed that such responses are associated with amelioration or resolution of infection. T cell responses in marmosets that may similarly correlate with protection from virus challenge have been detected in the NS3 and NS4A proteins of GB virus-B (Woollard et al., 2008).
Species demarcation criteria
Species in the Hepacivirus genus have been assigned based on their members’ genetic divergence, where viruses showing greater than 0.25 amino acid p-distances in a conserved region of NS3 (positions 1,123–1,566 as numbered in the HCV genotype 1a H77 reference sequence AF011751), and greater than 0.3 in the NS5B region (amino acid positions 2536–2959) are considered to be members of different species (Smith et al., 2016). Members of different hepacivirus species show characteristically restricted host ranges, typically infecting different host species. There are, however, a few exceptions; the bank vole may be infected by members of both species Hepacivirus myodae and Hepacivirus glareoli.
This demarcation point places hepaciviruses infecting humans and horses into different species (Hepacivirus hominis and Hepacivirus equi, respectively) while assigning the 7 relatively divergent genotypes of HCV described to date (Smith et al., 2014) to a single species. These HCV genotypes differ from each other by about 30–35% at the nt level (Simmonds et al., 2005). Within each genotype, there are a number of subtypes, differing from each other by about 15–25% at the nt level. Although genotypes are distinct genetically, discrimination of subtypes is less clear, particularly in areas of high diversity such as sub-Saharan Africa and Southeast Asia. Because systematic serological typing by virus neutralization has not been performed to date, and because major genotypes do not have any other taxonomic characteristics except, in some cases, geographic distribution and differences in treatment response, it was considered appropriate to classify the seven genotypes of HCV as members of same species (Hepacivirus hominis).
Related, unclassified viruses
|
Virus name |
Accession number |
Virus abbreviation |
|
Wenling shark virus |
WLSV-MHS-2 |
Virus names and virus abbreviations are not official ICTV designations.