Family: Anelloviridae
Simona Kraberger, Tanja Opriessnig, Fabrizio Maggi, Vladimir Celer, Hiroaki Okamoto, Lia van der Hoek, Philippe Biagini, Mart Krupovic and Arvind Varsani
The citation for this ICTV Report chapter is the summary to be published as Kraberger et al., (2026):
ICTV Virus Taxonomy Profile: Anelloviridae 2026, Journal of General Virology, (in press)
Corresponding author: Simona Kraberger ([email protected])
Edited by: Arvind Varsani and Evelien Adriaenssens
Posted: December 2025
Summary
Anelloviridae is a family of non-enveloped viruses with circular, negative-sense, single-stranded DNA genomes of 1.7–3.9 kb (Table 1 Anelloviridae). In the phylum Commensaviricota, this family includes 37 genera and 243 species. Anelloviruses have been identified in a broad range of mammals and birds, but are not associated with pathogenicity, except for members of the genus Gyrovirus.
Table 1 Anelloviridae. Characteristics of members of the family Anelloviridae
| Characteristic | Description |
| Example | torque teno virus 1 (AB041007), species Alphatorquevirus homin1 |
| Virion | Non-enveloped, icosahedral (T=1) capsid, 30–32 nm in diameter |
| Genome | Circular molecule of negative-sense ssDNA |
| Replication | Nuclear |
| Translation | From multiple, spliced mRNAs |
| Host range | Mammals, birds |
| Taxonomy | Realm Monodnaviria, kingdom Shotokuvirae, phylum Commensaviricota, class Cardeaviricetes, order Sanitavirales: 37 genera, 243 species |
Virion
Morphology
Anellovirids form non-enveloped capsids, 30–32 nm in diameter. The capsid comprises 60 copies of ORF1 protein that assemble into an icosahedral shell with the T=1 symmetry. The five-fold axes of symmetry display crown-like protrusions that are formed by the spike domains of the ORF1 protein (Liou et al., 2024) (Figure 1 Anelloviridae).
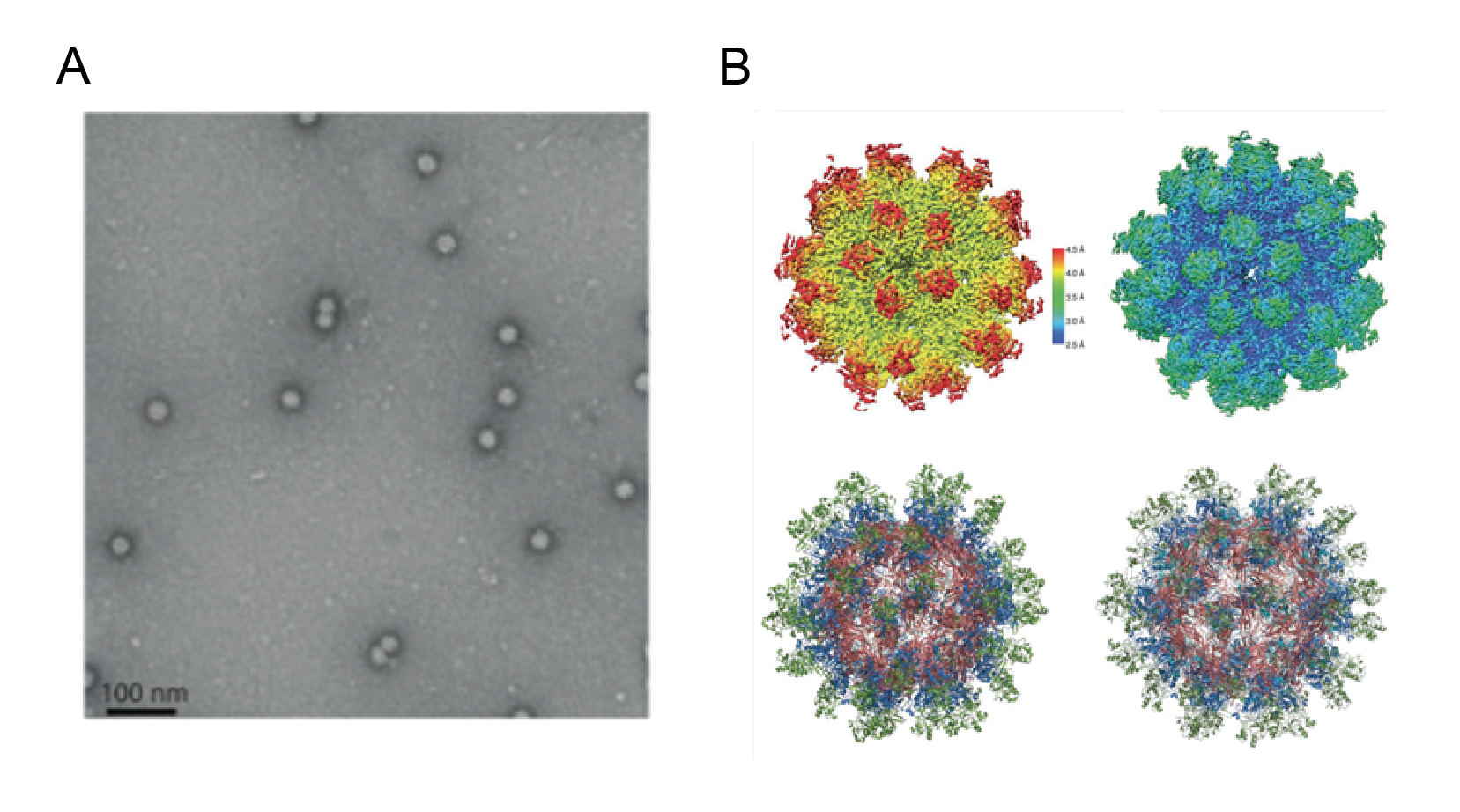 |
| Figure 1 Anelloviridae. Anellovirus particle. A. negative-stained electron microscopy image of anellovirus-like particles derived from TTMV-LY1 constructs. B. 3D reconstruction of anellovirus-like particles generated by the TTMV-LY1 ΔARM construct. Images courtesy of Ring Theraputics, Inc. from (Liou et al., 2024). |
Physicochemical and physical properties
The buoyant density of torque teno virus virions in CsCl is 1.31–1.33 g cm−3 as estimated using virions purified from serum (Mushahwar et al., 1999).
Nucleic acid
The genome of anelloviruses is a single negative-sense circular ssDNA molecule, of 1.7–3.9 kb. The putative non-coding region generally contains a tract of high G+C content, which is postulated to form a secondary structure composed of stem loops.
Proteins
Anelloviruses generally encode three main proteins known as orf1 (vp1 in gyroviruses), orf2 (vp2), and orf3 (vp3); additional open reading frames are present in some species. The minor ORFs are typically overlapping with ORF1, which spans a large portion of the genome. Orf1 encodes the capsid protein, the N-terminal region of which contains an arginine-rich hydrophilic region postulated to be important for nuclear localization, signaling and binding to the viral genome (Petersen et al., 2025). Additionally, for anelloviruses with larger genomes, the C-terminal region of the capsid also carries a nuclear localization signal (Petersen et al., 2025). The ORF1 proteins of all anelloviruses adopt a conserved jelly-roll domain, which plays a key role in the capsid assembly. In many members, the jelly-roll domain contains a spike domain insertion, comprising subdomains P1 and P2 (Liou et al., 2024). The spike domains protrude away from the capsid surface, forming crown-like structures at the five-fold symmetry axes. Depending on the size of the spike domain insertion as well as N- and C-terminal extensions, ORF1 capsid proteins can vary in size from approximately 340 to 1020 amino acids, with the largest ORF1 proteins thus far having been identified in anelloviruses from oceanic dolphins in the Delphinidae family (De Koch et al., 2025).
Genome organization and replication
Genomes of anelloviruses contain at least three main open reading frames, orf1, orf2 and orf3, with additional open reading frames in some species (Figure 2 Anelloviridae). The proportion of the genome occupied by orf1 also varies widely between species (De Koch et al., 2025). The intergenic region contains a GC-rich region. Orf1 encodes a capsid protein. There is evidence that the proteins encoded by orf2 and orf3 play a role in initiating replication, and it has been hypothesized the genome replication occurs is through a recombination-dependent mechanism with the aid of host replication factors (Boisvert et al., 2025).
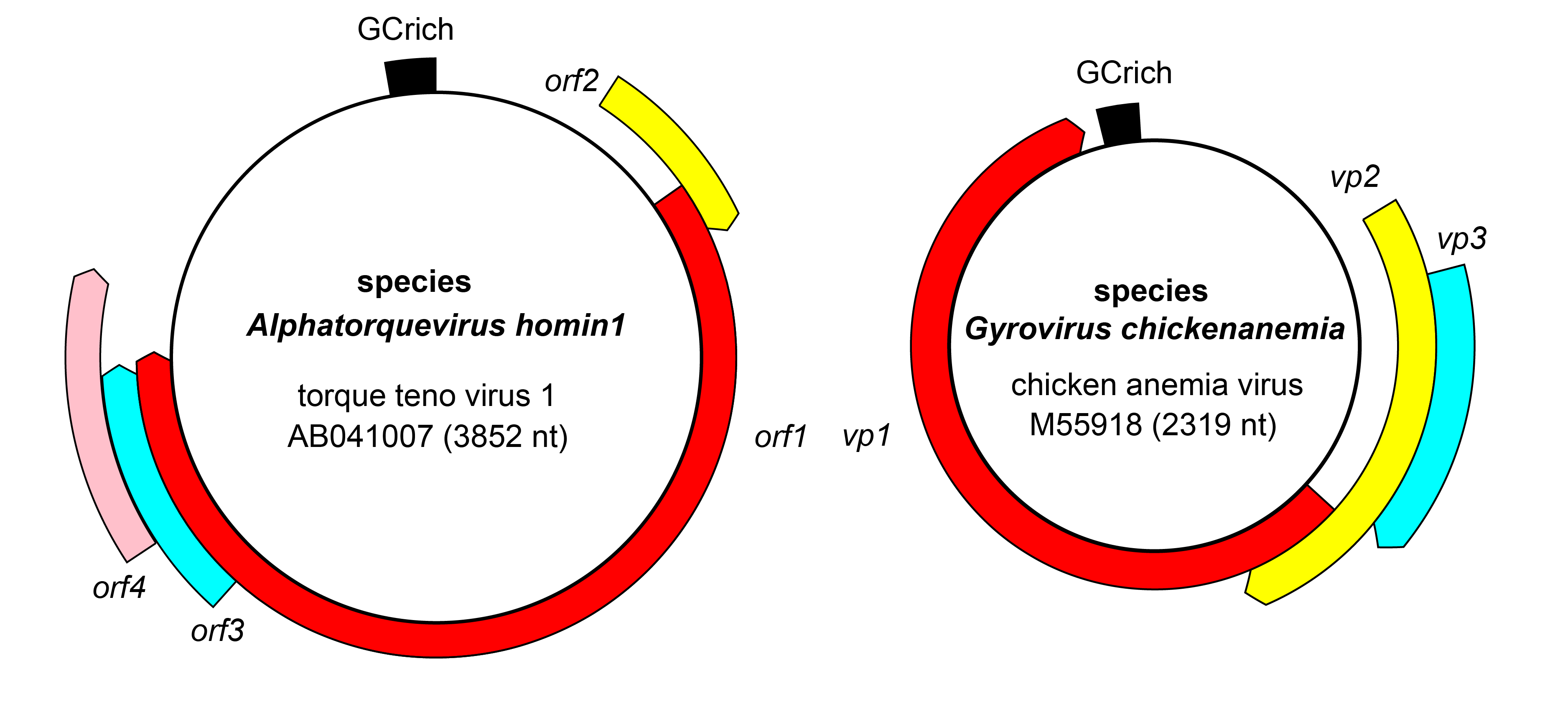 |
| Figure 2 Anelloviridae. Genome organization of representative anelloviruses. |
Biology
Anelloviruses are genetically diverse and co-infections are often detected in a single host. They are highly prevalent in human populations globally and have also been identified in a broad range of other mammals and birds. Most members of the genus Gyrovirus have been recovered from birds, supporting the view that members of this genus primarily infect avian species; detection of gyroviruses in mammalian fecal samples are thought to be diet-derived. The remaining 36 genera include anelloviruses which were largely identified in various samples of mammals; the few recovered from blood-feeding invertebrates are probably blood-meal derived.
Direct links to disease have not been established for anelloviruses apart from those in the genus Gyrovirus. The prototypical gyrovirus, chicken anemia virus, is associated with immunosuppressive disease in chickens resulting in a range of clinical outcomes including weight loss, anaemia and intramuscular haemorrhaging (Fatoba and Adeleke 2019). For other anelloviruses, their wide distribution across mammalian host species and populations, and lack of disease links, has led to the hypothesis that they are commensals (Kaczorowska and van der Hoek 2020).
Antigenicity
Particles of torque teno virus in blood are bound to immunoglobulins G (IgG) and M (IgM), forming immune complexes (Itoh et al., 2000, Tsuda et al., 2001).
Derivation of names
Aleptorquevirus: from alep, the first letter of the Phoenician alphabet
Alphatorquevirus: after alpha, the first letter of the Greek alphabet
Anelloviridae: from the Latin anello, meaning "ring", referring to the circular nature of the DNA genome of viruses in the family; the suffix ‑viridae for family taxa
Ayintorquevirus: after ayin, the sixteenth letter of the Phoenician alphabet
Betatorquevirus: after beta, the second letter of the Greek alphabet
Chitorquevirus: after chi, the twenty second letter of the Greek alphabet
Deltatorquevirus: after delta, the fourth letter of the Greek alphabet
Epsilontorquevirus: after epsilon, the fifth letter of the Greek alphabet
Etatorquevirus: after eta, the seventh letter of the Greek alphabet
Gammatorquevirus: after gamma, the third letter of the Greek alphabet
Gimeltorquevirus: after gimel, the third letter of the Phoenician alphabet
Gyrovirus: after gyros, the Greek word for round or circular
Hetorquevirus: after he, the fifth letter of the Phoenician alphabet
Iotatorquevirus: after iota, the ninth letter of the Greek alphabet
Kappatorquevirus: after kappa, the tenth letter of the Greek alphabet
Lambdatorquevirus: after lambda, the eleventh letter of the Greek alphabet
Lamedtorquevirus: after lamed, the twelfth letter of the Phoenician alphabet
Memtorquevirus: after mem, the thirteenth letter of the Phoenician alphabet
Mutorquevirus: after mu, the twelfth letter of the Greek alphabet
Nutorquevirus: after nu, the thirteenth letter of the Greek alphabet
Omegatorquevirus: after omega, the twenty fourth letter of the Greek alphabet
Omicrontorquevirus: after omicron, the fifteenth letter of the Greek alphabet
Petorquevirus: after pe, the seventeenth letter of the Phoenician alphabet
Pitorquevirus: after pi, the sixteenth letter of the Greek alphabet
Psitorquevirus: after psi, the twenty third letter of the Greek alphabet
Qoptorquevirus: after qop, the nineteenth letter of the Phoenician alphabet
Rhotorquevirus: after rho, the seventeenth letter of the Greek alphabet
Sadetorquevirus: after sade, the eighteenth letter of the Phoenician alphabet
Samektorquevirus: after samek, the fifteenth letter of the Phoenician alphabet
Sigmatorquevirus: after sigma, the eighteenth letter of the Greek alphabet
Tettorquevirus: after tet, the ninth letter of the Phoenician alphabet
Thetatorquevirus: after theta, the eighth letter of the Greek alphabet
Upsilontorquevirus: after upsilono, the twentieth letter of the Greek alphabet
Wawtorquevirus: after waw, the sixth letter of the Phoenician alphabet
Xitorquevirus: after xi, the fourteenth letter of the Greek alphabet
Yodtorquevirus: after yod, the tenth letter of the Phoenician alphabet
Zayintorquevirus: after zayin, the seventh letter of the Phoenician alphabet
Zetatorquevirus: after zeta, the sixth letter of the Greek alphabet
Demarcation criteria for taxa within the family
Genus-level demarcation criteria are based on phylogenetic analyses using the ORF1 amino acid sequence, and species-level demarcation is based on comparisons of the orf1 gene sequences with members of different species having a pairwise identity of <69%.
Relationships within the family
Anelloviridae is a highly diverse family of viruses. Members of the genus Gyrovirus form an outlier clade, while members of the other genera form distinct subclades (Figure 3 Anelloviridae).
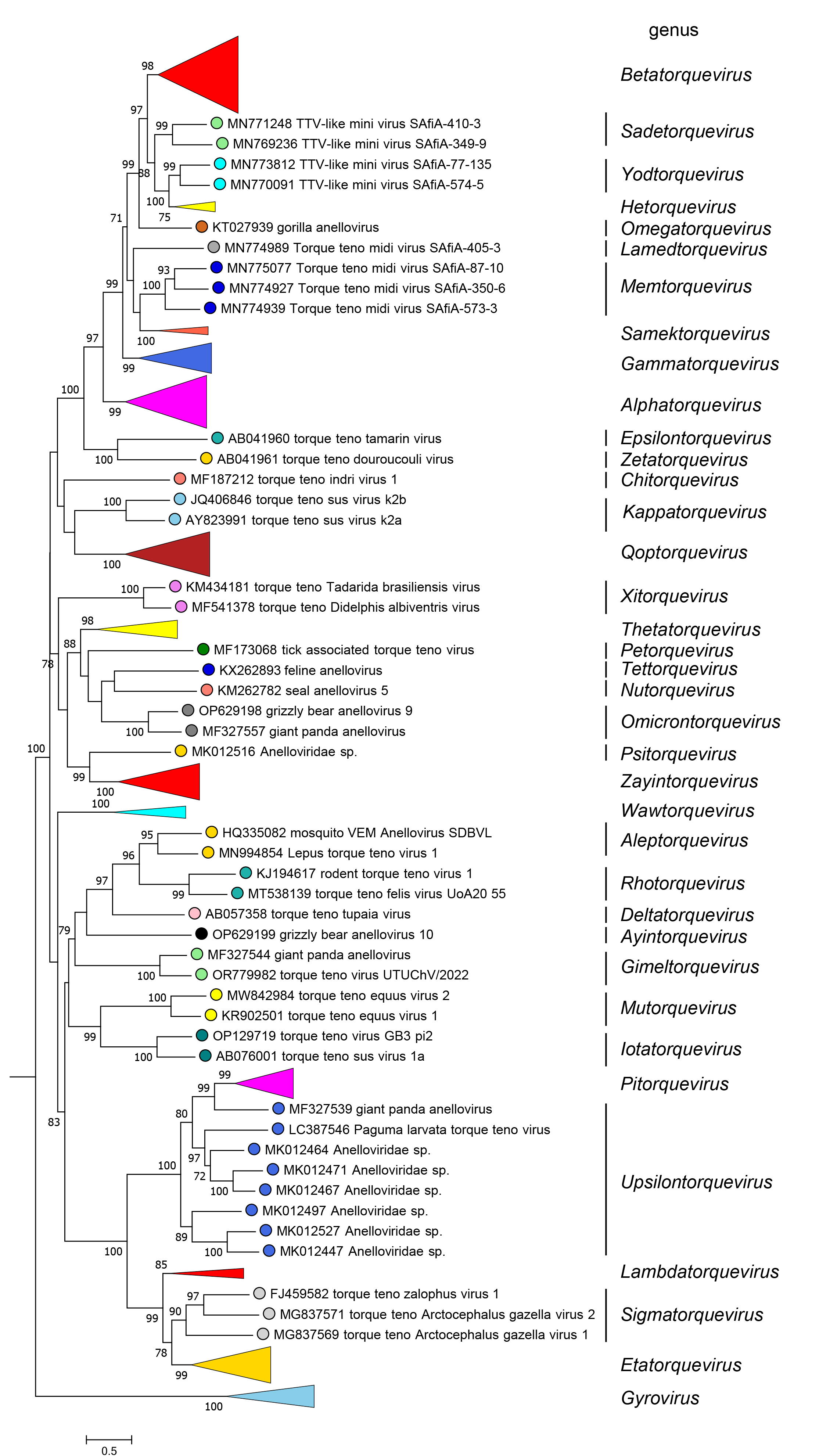 |
| Figure 3 Anelloviridae. Condensed maximum-likelihood phylogenetic tree of ORF1 protein sequences of members of the family Anelloviridae comprising representatives for each species and collapsed branches for genera with more than three species (apart from the polyphyletic genus Upsilontorquevirus). Sequences are identified on the figure by their GenBank accession numbers. The phylogenetic tree was rooted with the gyroviruses. The full tree, including all accession numbers and virus names, is available on a separate page. |
Relationships with other taxa
Due to the absence of a typical replication initiation protein found in other eukaryotic ssDNA viruses and distinct genomic features, Anelloviridae is the sole family in the phylum Commensaviricota. However, anelloviruses are believed to be evolutionarily related to members of the phylum Cressdnaviriota, in particular, to viruses within the family Circoviridae (Butkovic et al., 2023). The relationship has been inferred based on the similarity of jelly-roll fold capsid proteins shared by anelloviruses and circoviruses. Thus, it has been suggested that anelloviruses have evolved from a circovirus-like ancestor (Butkovic et al., 2023).

