Family: Solemoviridae
Genus: Enamovirus
Distinguishing features
Enamoviruses have a polycistronic, positive-sense RNA genome that consists of non-coding 5′- and 3′-regions and five ORFs. ORFs 0, 1 and 2 are all translated from the genomic RNA (Figure 1. Enamovirus). Translation of ORF1 occurs via a leaky scanning mechanism. The RNA-directed RNA polymerase (RdRP) encoded by ORF2 is expressed as a fusion protein through a −1 ribosomal frameshift mechanism. The 3′-proximal ORFs ORF3 and ORF5 are translated from the subgenomic RNA. ORF5 is only expressed as a fusion protein via a readthrough of the ORF3 stop codon. Enamoviruses lack ORF4 and are known to depend on the symbiotic viruses belonging to genus Umbravirus (family Tombusviridae) for efficient movement in the plant (Demler et al., 1994).
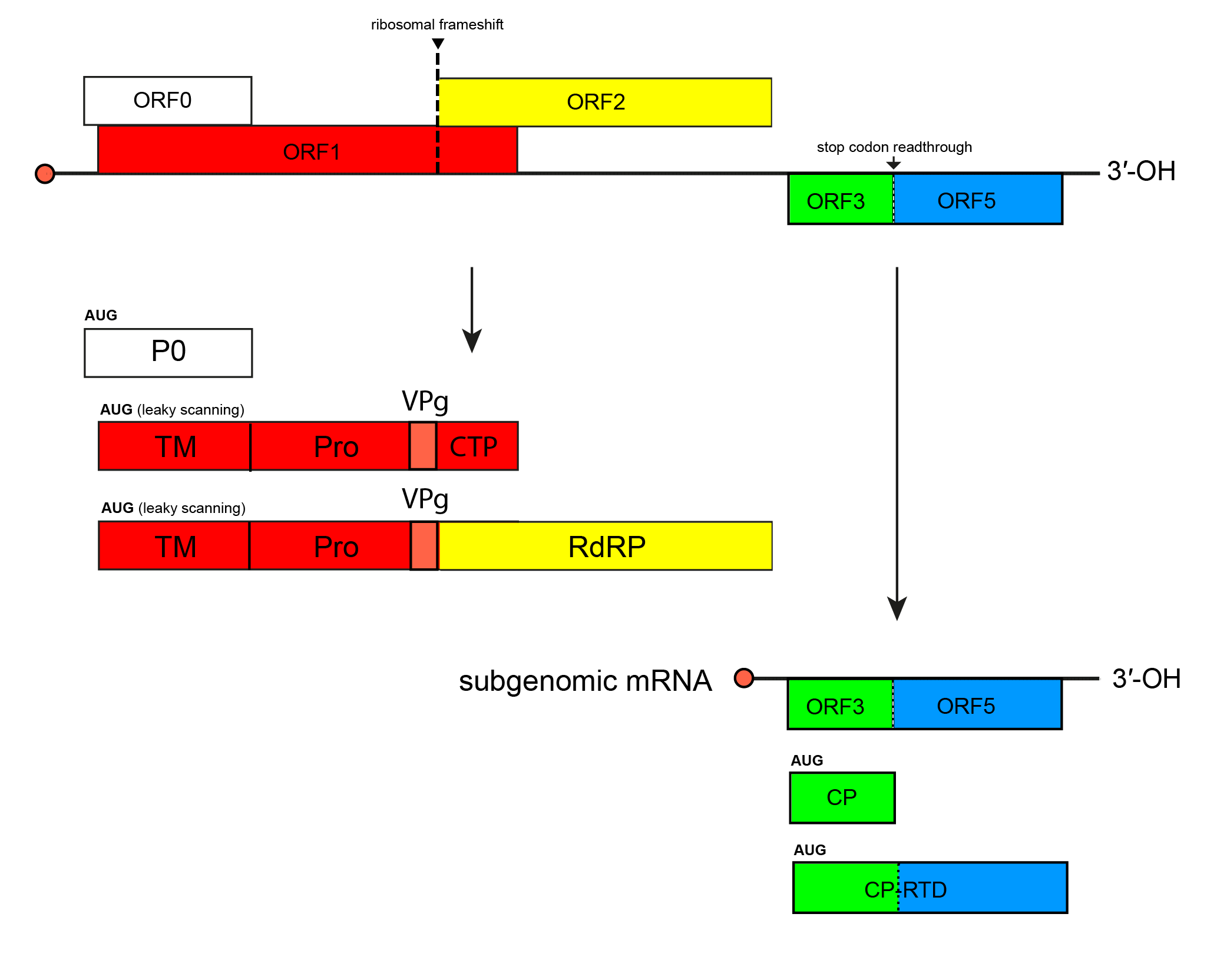 |
| Figure 1. Enamovirus Transcriptional and translational map of the genome of pea enation mosaic virus 1 (genus Enamovirus). Open reading frames (ORFs) are indicated by their position above and below the genome in the order -1, 0 and +1; those with sequence homology at the family level are represented by coloured boxes, while those with no sequence homology are shown colourless. P0, protein P0; TM, transmembrane domain; Pro, serine protease domain; VPg, virus protein genome-linked; CTP, C-terminal domain; RdRP, RNA-directed RNA polymerase; CP, coat protein; RTD, readthrough domain. |
Virion
Morphology
See discussion under family description.
Physicochemical and physical properties
Enamovirus virions have a Mr of about 5.6×106, buoyant densities in CsCl of 1.42 g cm−3, and S20,w of 107–122S.
Nucleic acid
The genome is 5,706 nt for pea enation mosaic virus 1 and 6,317 nt for grapevine enamovirus 1. See further discussion under family description.
Proteins
See discussion under family description.
Genome organization and replication
See discussion under family description.
Biology
Enamoviruses are transmitted by aphids in persistent, circulative and non-propagative manner. A gut membrane alanyl aminopeptidase N has been identified as a receptor for pea enation mosaic virus 1 (PEMV1) in the aphid vector Acyrthosiphon pisum Harris, 1776 (Linz et al., 2015). Studies of PEMV1 indicate it is present as part of a complex with the symbiotic umbravirus pea enation mosaic virus 2 (PEMV2) and induces mosaic symptoms and enations. PEMV1 is readily transmitted mechanically, a property dependent on its multiplication in cells co-infected with PEMV2. Whereas PEMV1 provides the capsid for both viruses, PEMV2 complements the lack of movement protein of PEMV1 and allows it to relieve phloem restriction (Doumayrou et al., 2016). Aphid transmissibility is conferred by PEMV1, but can be lost after several mechanical passages. Virions are found in mesophyll tissue as well as in vascular tissue. The genome of PEMV1 is capable of autonomous replication, but is dependent on PEMV2 to support systemic invasion. However, several recently discovered enamoviruses lack coinfection with umbraviruses and may be associated with nucleorhabdoviruses (Cao et al., 2019, Bejerman et al., 2016, Debat and Bejerman 2019).
Species demarcation criteria
Criteria used to demarcate species of the genus include:
- Differences in breadth and specificity of host range
- Failure of cross-protection in either one-way or two-way relationships
- Differences in serological specificity with discriminatory polyclonal or monoclonal antibodies
- Differences in amino acid sequence identity of any gene product of greater than 10%.

