Family: Baculoviridae
Genus: Betabaculovirus
Distinguishing features
Occlusion bodies (OBs) are generally ovocylindrical in shape, measure approximately 0.12 × 0.50 µm, and characteristically contain one virion (Tanada and Hess 1991) (Figure 1.Betabaculovirus). Each occlusion-derived virus virion typically contains a single nucleocapsid within a single envelope. Occluded virions may mature among nuclear-cytoplasmic cellular contents after loss of the nuclear membrane of infected cells. Uncoating is thought to occur by a mechanism in which viral DNA is extruded into the nucleus through the nuclear pore while the capsid remains in the cytoplasm (Summers 1971, Au et al., 2013).
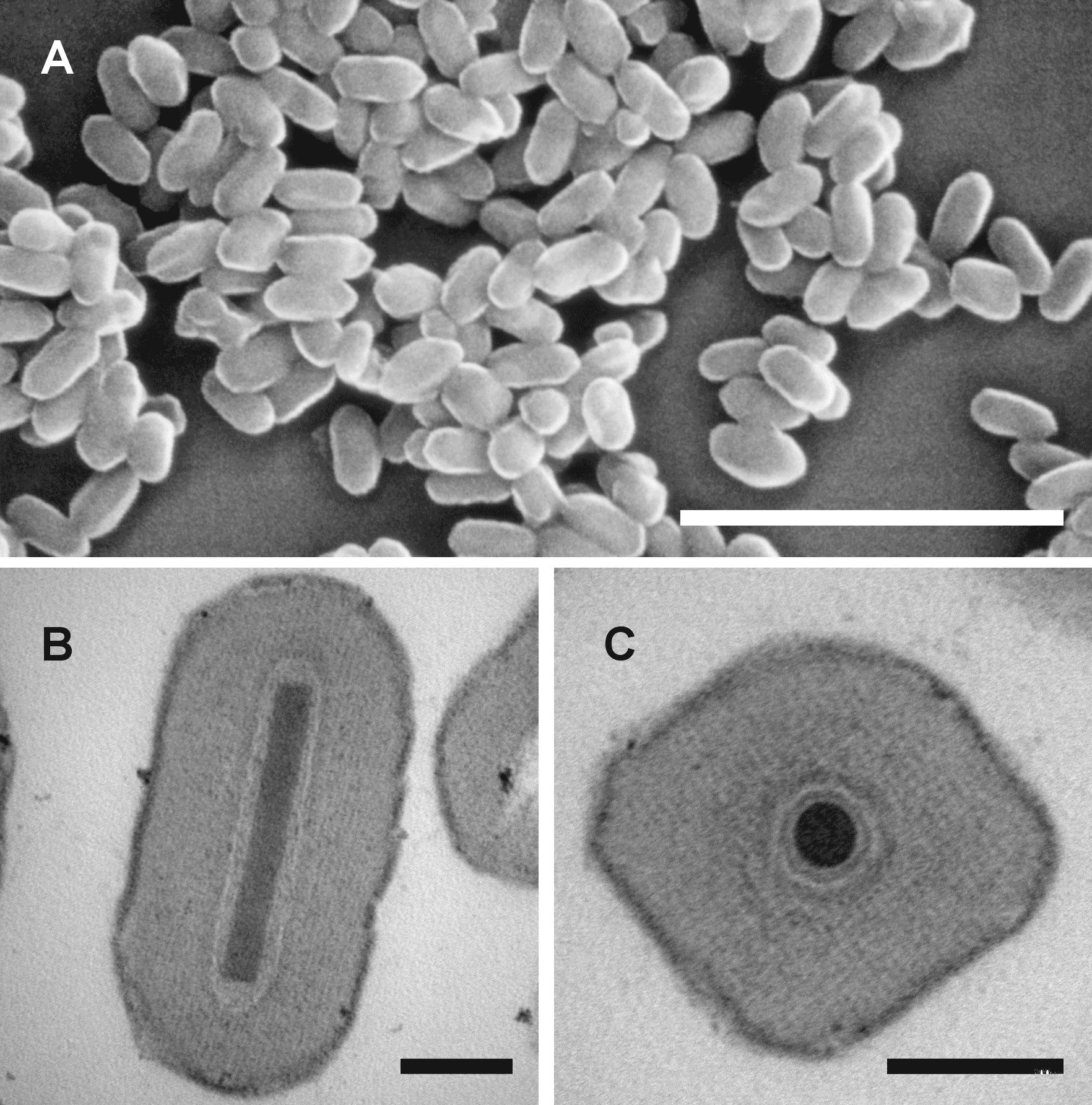 |
| Figure 1. Betabaculovirus. Occlusion bodies of baculoviruses in the genus Betabaculovirus. Scanning electron micrograph of Spodoptera frugiperda granulovirus occlusion bodies (A, courtesy of J. R. Adams), and transmission electron micrographs of Mythimna unipuncta granulovirus B occlusion bodies (B and C), showing their unique ovocylindrical shape. Scale bars: (A) 2 µm; (B, C) 100 nm. |
Virion
See discussion under family properties.
Genome organization and replication
See discussion under family properties.
Biology
Members of this genus have been isolated only from the insect order Lepidoptera.
Species demarcation criteria
Traditionally, species distinctions have been broadly based on host range and specificity, DNA restriction profiles, DNA sequences from various regions of the genome, and predicted protein sequence similarities. More recently, species demarcation criteria for alpha- and betabaculoviruses have been proposed that rely upon pairwise nucleotide distances estimated with the Kimura-2-parameter substitution model from partial sequences of three conserved baculovirus genes: late expression factor-8 (lef-8) and late expression factor-9 (lef-9), which encode subunits of the baculovirus RNA polymerase, and polyhedrin/granulin (polh/gran), which encodes the viral occlusion body matrix protein (Jehle et al., 2006b). If nucleotide distances between two viruses at these loci are less than 0.015 substitutions/site, the two baculoviruses are considered to belong to the same species. If nucleotide distances between two viruses are greater than 0.05 substitutions/site, the viruses are considered to belong to different species. If the nucleotide distances lie between 0.015 and 0.050 substitutions/site, additional characteristics of the two viruses (i.e. host range) must be considered to make a decision about their taxonomic status. The proposed criteria were originally based on an alignment of sequences from 117 separate baculovirus isolates and the phylogeny inferred from this alignment. Researchers have applied this criterion to other isolates to identify many new baculovirus species and variants of currently recognized species. A more recent examination of pairwise nucleotide distances for all 38 baculovirus core genes among 172 completely sequenced baculovirus genomes has confirmed the current species classification based on pairwise distances for lef-8, lef-9, and polh loci (Wennmann et al., 2018).

