Family: Solemoviridae
Genus: Polerovirus
Distinguishing features
Poleroviruses have a polycistronic, positive-sense, RNA genome that consists of non-coding 5′- and 3′-regions and 7 ORFs, four of which (ORF3a, ORF4, ORF6 and ORF7) are unique to poleroviruses. ORFs 0, 1 and 2 are all translated from the genomic RNA (Figure 1. Polerovirus). Translation of ORF1 occurs via a leaky scanning mechanism. The RNA-directed RNA polymerase (RdRP) encoded by ORF2 is expressed as a fusion protein through a −1 ribosomal frameshift mechanism. The 3′-proximal ORFs 3a, 3, 4 and 5 are translated from subgenomic RNA1, while ORF6 and ORF7 are translated from subgenomic RNA2; ORF7 is also translated from subgenomic RNA3. ORF5 is only expressed as a fusion protein via a readthrough of ORF3 stop codon. In addition, the genome of potato leafroll virus (PLRV) encodes a replication-associated protein Rap1 from its unique ORF8.
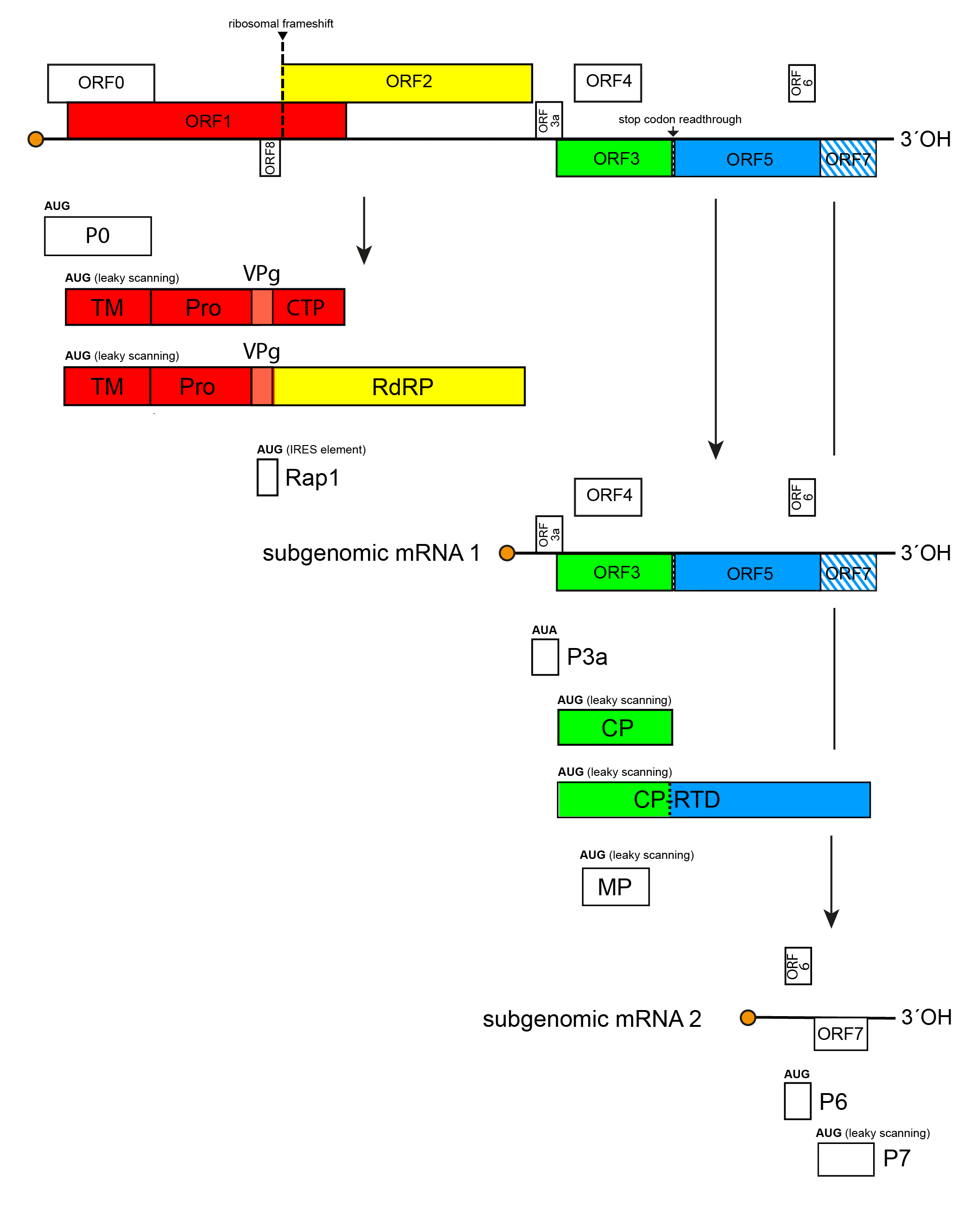 |
| Figure 1. Polerovirus Transcriptional and translational map of the genome of potato leafroll virus (genus Polerovirus). Open reading frames (ORFs) are indicated by their position above the genome in the order -1, 0 and +1; those with sequence homology at the family level are represented by coloured boxes, while those with no sequence homology are shown colourless. Hatched area indicates overlap between the ORFs. P0, protein P0; Rap1, replication-associated protein 1; TM, transmembrane domain; Pro, serine protease domain; VPg, virus protein genome-linked; CTP, C-terminal domain; RdRP, RNA-directed RNA polymerase; P3a, protein 3a; MP, movement protein; CP, coat protein; RTD, readthrough domain, P6, protein P6; P7, protein P7. ORF7 is expressed from both sgRNA2 and sgRNA3 (not shown). |
Virion
Morphology
See discussion under family description.
Physicochemical and physical properties
Virion buoyant density in CsCl is 1.39–1.42 g cm−3; S20,w is 115–127S.
Nucleic acid
Genome ranges from 5,625 nt for maize yellow mosaic virus and 6,244 nt for pepper vein yellows virus 1, pepper vein yellows virus 3 and pepper vein yellows virus 4. See further discussion under family description.
Proteins
See discussion under family description.
Genome organization and replication
See discussion under family description.
Biology
Polerovirus infections are phloem-specific and transmitted by aphid vectors. Infection is often characterized by phloem necrosis, streaking, yellowing, rolling and thickening of the leaves and stunting of the plants. Viruses are acquired by vector phloem feeding, the viruses entering the hemocoel of the aphid gut by a receptor-mediated transport process, following which they circulate in the hemolymph and then enter the accessory salivary gland by a second receptor-mediated transport event. Movement across tissue barriers within the gut and accessory salivary glands occurs via clathrin-mediated endocytosis (Gray and Gildow 2003). The ephrin receptor protein of the aphid Myzus persicae (Sulzer, 1776) has been identified as a putative receptor for the turnip yellows virus, beet mild yellowing virus, and cucurbit aphid borne yellows virus (Mulot et al., 2018). Cyclophilin B has been implicated in the transmission of cereal yellow dwarf virus RPV by the aphid Schizaphis graminum (Rondani, 1852) (Tamborindeguy et al., 2013). The unassigned polerovirus, pepper whitefly-borne vein yellows virus, is transmitted by the whitefly Bemisia tabaci (Gennadius, 1889) (Ghosh et al., 2019).
Several studies describe polerovirus coinfections with other viruses (Moreno and López-Moya 2020). Interestingly, a number of umbraviruses depend on assistance from poleroviruses but this relationship is not obligatory for the assistor poleroviruses as in the case of coinfection with the umbravirus pea enation mosaic virus (PEMV2) and the assister enamovirus pea enation mosaic virus 1 (PEMV1) (Zhou et al., 2017, Ryabov et al., 2001).
Species demarcation criteria
Criteria used to demarcate species of the genus include:
- Differences in breadth and specificity of host range
- Failure of cross-protection in either one-way or two-way relationships
- Differences in serological specificity with discriminatory polyclonal or monoclonal antibodies
- Differences in amino acid sequence identity of any gene product of greater than 10%
- Differences in nucleotide sequence identity of genomes around or greater than 25%
Member species
The Member Species table enumerating important virus exemplars classified under each species of the genus is provided at the bottom of the page.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Allium polerovirus A | MH898527 | APVA |
| cotton bunchy top virus | JF803842 | CBTV |
| cucumber aphid-borne virus | FJ460218 | CuABYV |
| fennel motley virus | LT595018 | FMV |
| lettuce yellows virus | LN865080 | LYV |
| Malva chlorosis virus | LT595029 | MChV |
| Setaria yellow dwarf virus | LC333290 | SYDV |
| spinach yellows virus | LN865083 | SYV |
| Suaeda polerovirus A | MH898529 | SPoA |
| tobacco virus 2 | KY038943 | TV2 |
| tobacco yellow virus | MK751486 | TYV |
| wild carrot motley leaf virus | LT595010 | WCMLV |
| wild carrot red leaf virus | LT615231 | WCRLV |
Virus names and virus abbreviations are not official ICTV designations.

