Family: Caliciviridae
Jan Vinjé, Mary K. Estes, Pedro Esteves, Kim Y. Green, Kazuhiko Katayama, Nick J. Knowles, Yvan L’Homme, Vito Martella, Harry Vennema and Peter A. White
The citation for this ICTV Report chapter is the summary published as Vinjé et al., (2019):
ICTV Virus Taxonomy Profile: Caliciviridae, Journal of General Virology, 100, 1469–1470
Corresponding author: Jan Vinjé ([email protected])
Edited by: Nick J. Knowles and Stuart G. Siddell
Posted: September 2019
PDF: ICTV_Caliciviridae.pdf
Summary
The family Caliciviridae includes viruses with a linear positive-sense RNA genome of 6.4–8.5 kb with the non-structural and structural proteins encoded by different ORFs (Table 1. Caliciviridae). Virions are non-enveloped particles ranging from 27–40 nm in diameter with an icosahedral symmetry. The family includes eleven genera of which members of seven infect mammals (Lagovirus, Norovirus, Nebovirus, Recovirus, Sapovirus, Valovirus, and Vesivirus), members of two genera infect birds (Bavovirus, Nacovirus), and members of two genera infect fish (Minovirus and Salovirus). Each genus includes 1–2 species. Human noroviruses are a leading cause of acute gastroenteritis in humans. Furthermore, unclassified caliciviruses have been detected in geese, yellowfin seabream, greater green snake, arctic lamprey, frogs and various Australian birds (Wang et al., 2017, Shi et al., 2018, Wille et al., 2018), highlighting the wide host range of viruses in the family Caliciviridae.
Table 1. Caliciviridae. Characteristics of members of the family Caliciviridae
| Characteristic | Description |
| Typical member | Norwalk virus (M87661), species Norovirus norwalkense |
| Virion | Non-enveloped with icosahedral symmetry, 27–40 nm in diameter |
| Genome | Linear positive-sense genomic RNA of 7.4–8.3 kb, with a 5′-terminal VPg and 3′-terminal poly(A) |
| Replication | Cytoplasmic |
| Translation | From genome-sized (non-structural proteins) and 3′-terminal subgenomic (structural proteins) RNAs |
| Host range | Mammals (Lagovirus, Norovirus, Nebovirus, Recovirus, Sapovirus Valovirus, and Vesivirus), birds (Bavovirus, Nacovirus), fish (Minovirus, Salovirus) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Picornavirales; eleven genera, each including one or two species |
Virion
Morphology
Calicivirus virions are non-enveloped approximately 35–40 nm in diameter with an icosahedral symmetry (Figure 1. Caliciviridae). The capsid is composed of 90 dimers of the major structural protein VP1 arranged on a T=3 icosahedral lattice (Prasad et al., 1999). In noroviruses, VP1 forms a subunit comprised of a shell (S) domain and two protruding (P) domains which consist of P1 and P2 subdomains (Figure 1. Caliciviridae). The P2 subdomain contains potential neutralizing antibody epitopes and interacts with histo-blood group antigens, which are a diverse family of carbohydrates that serve as binding ligands for virus entry. Characteristically, calicivirus capsid architecture has 32 cup-shaped depressions at each of the icosahedral five-fold and three-fold axes. In negatively-stained virus preparations, some cup-shaped depressions appear distinct and well defined, while in others these depressions are less prominent.
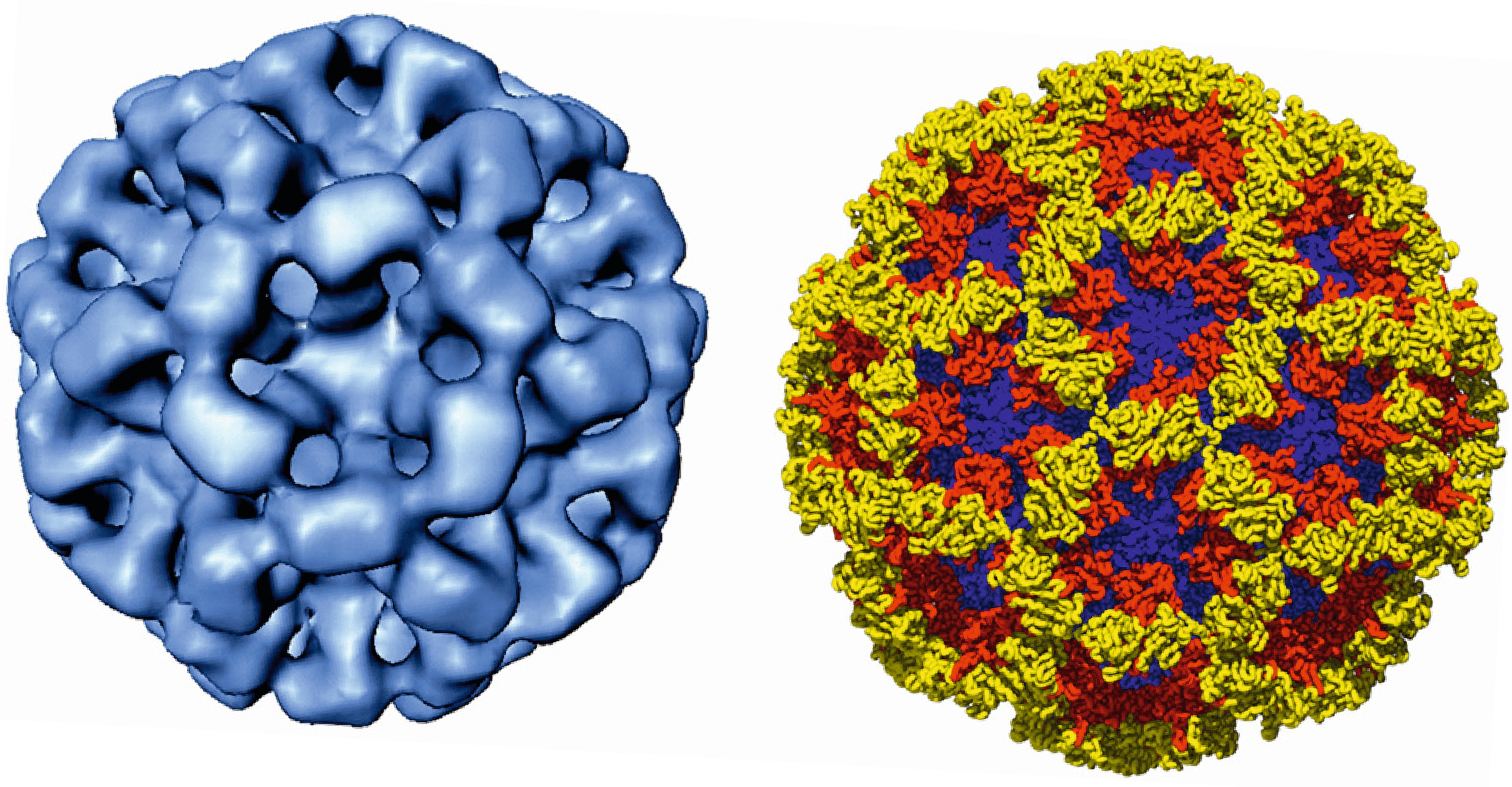 |
| Figure 1. Caliciviridae. The structure of the calicivirus capsid exemplified by cryo-image reconstruction of recombinant Norwalk virus -like particles (Left). X-ray structure of the Norwalk virus capsid (Right) with the shell, protruding 1, and protruding 2 domains colored in blue, red and yellow, respectively. (Courtesy of B.V. Prasad.) |
Physicochemical and physical properties
Virion Mr is about 15×106. Virion buoyant density is 1.33–1.41 g cm−3 in CsCl and 1.29 g cm−3 in glycerol-potassium tartrate gradients. Virion S20,w is 170–187S. Physicochemical properties have been established for some members of the family. Generally, caliciviruses are stable in the environment and many strains are resistant to inactivation by heat and certain chemicals (ether, chloroform and mild detergents). Enteric caliciviruses are acid-stable.
Nucleic acid
The calicivirus genome consists of a linear, positive sense, ssRNA molecule of 7.4–8.3 kb with VPg (virus protein genome-linked) covalently bound to the 5′-end and a 3′-poly(A)-tail. A subgenomic RNA (sgRNA) of 2.2–2.4 kb is synthesized intracellularly and is VPg-linked (Asanaka et al., 2005).
Proteins
Virions are predominantly comprised from one major capsid protein, VP1 (58–60 kDa), a minor structural protein, VP2 (8.5–23 kDa) and VPg (13–16 kDa). Each virion consist of 180 copies of VP1 (90 dimers), one or two copies of both VP2 and VPg (Sosnovtsev and Green 2000). The expression of VP2 is believed to occur by termination codon-dependent reinitiation translation (Napthine et al., 2009). VPg, which is essential for viral replication, is covalently attached to the 5′-terminus of the genomic and subgenomic RNA (Figure 2. Caliciviridae) and requires interaction with eukaryotic initiation factors (eIF3, eIF4E, eIF4G), further supporting a role in translation. VPg and other non-structural proteins map to genomic regions analogous to the non-structural domains of picornaviruses (Figure 2. Caliciviridae).
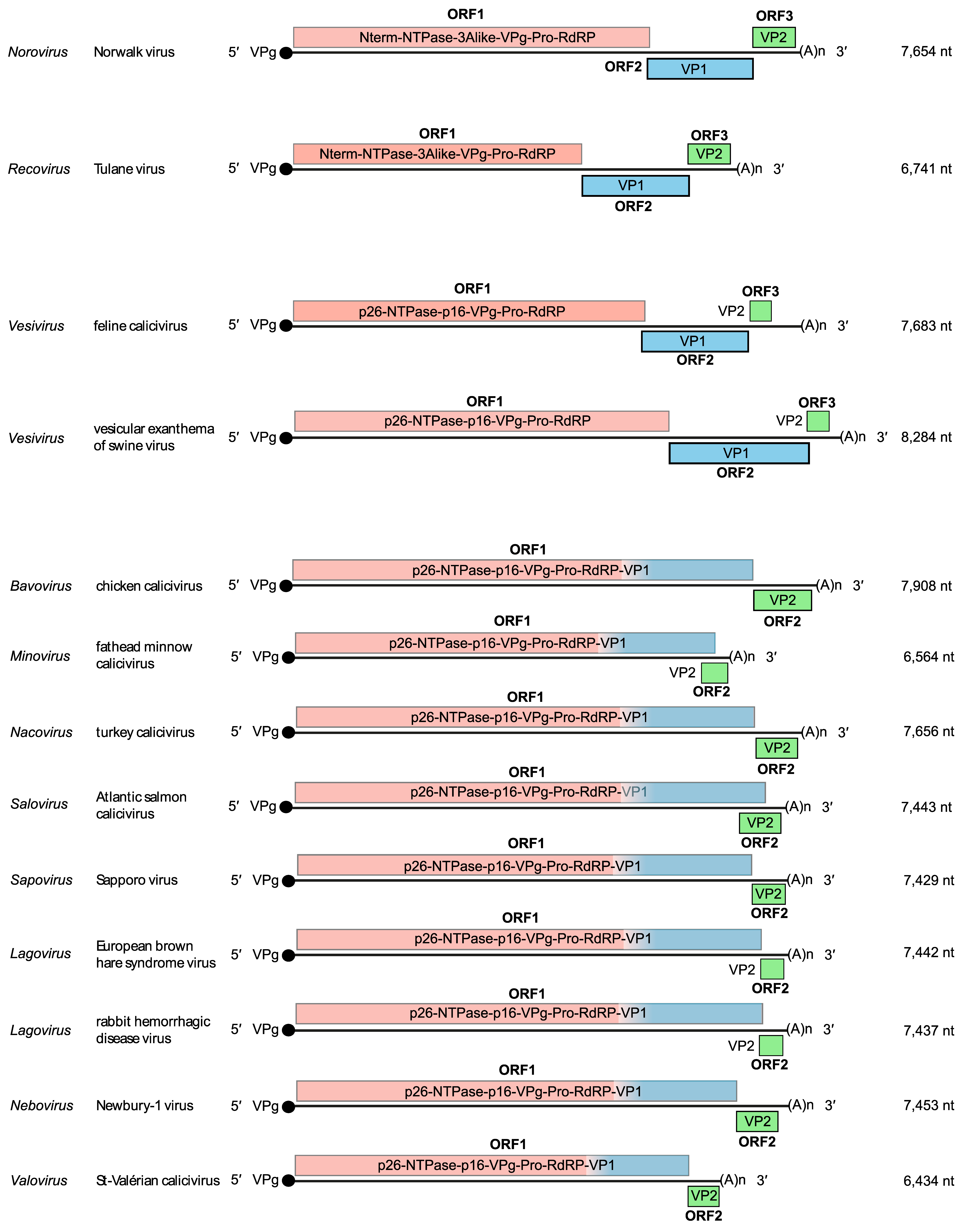 |
| Figure 2. Caliciviridae. Diagrammatic representation of open reading frame usage (ORF) and gene order in the family Caliciviridae. Caliciviruses have a positive-strand RNA genome (7.4–8.3 kb), which is covalently linked at the 5′-end to a protein (VPg, depicted by a black circle). The full length genome (of members of the genera indicated) is organized into 2–4 ORFs. ORF1 encodes a polyprotein that is posttranslationally-cleaved into individual non-structural proteins. Cleavage sites have been described for several genera but there is considerable variation in lengths of the proteins. |
Lipids
Although enteric caliciviruses are shed in stool samples as non-enveloped virus, there is evidence that human norovirus is shed enclosed within vesicles of exosomal or plasma membrane origin. These vesicles remain intact during fecal-oral transmission and thereby transport multiple viral particles, which may enhance disease severity (Santiana et al., 2018).
Carbohydrates
None reported.
Genome organization and replication
The virion RNA of caliciviruses is infectious and its genome acts as an mRNA template comprised of two open reading frames (ORFs) for members of the genera Bavovirus, Lagovirus, Minovirus, Nacovirus, Nebovirus, Salovirus and Sapovirus, and three ORFs for members of the genera Norovirus, Recovirus, Vesivirus and Valovirus (Figure 2. Caliciviridae). Documented exceptions include murine noroviruses which possess a fourth ORF (ORF4) that encodes a virulence factor (VF1) (McFadden et al., 2011) (Figure 3. Caliciviridae) and feline calicivirus (FCV) which encodes a leader capsid (LC) that is proteolytically processed by the viral protease to yield the mature capsid protein (Sosnovtsev et al., 1998). The 5′-proximal ORF1 encodes a large polyprotein which is co- and post-translationally cleaved by the virus-encoded protease (NS6pro) into at least six mature nonstructural proteins (NS1/2, NS3, NS4, NS5, NS6, and NS7) (Karst et al., 2014), which are expressed in the same order for members of each genus (Figure 2. Caliciviridae). For bavoviruses, lagoviruses, minoviruses, nacoviruses, neboviruses, saloviruses, and sapoviruses, ORF1 encoding the non-structural proteins is in-frame and contiguous with VP1. A 5′-end VPg-linked subgenomic RNA is produced intracellularly during translation and drives the initiation of translation and produces VP1 and VP2 (Anasaka 2005) (Figure 3. Caliciviridae). A dsRNA genome corresponding to full-length genomic ssRNA has been identified in FCV and San Miguel sea lion virus-infected cells, indicating that replication occurs via a negative-strand intermediate.
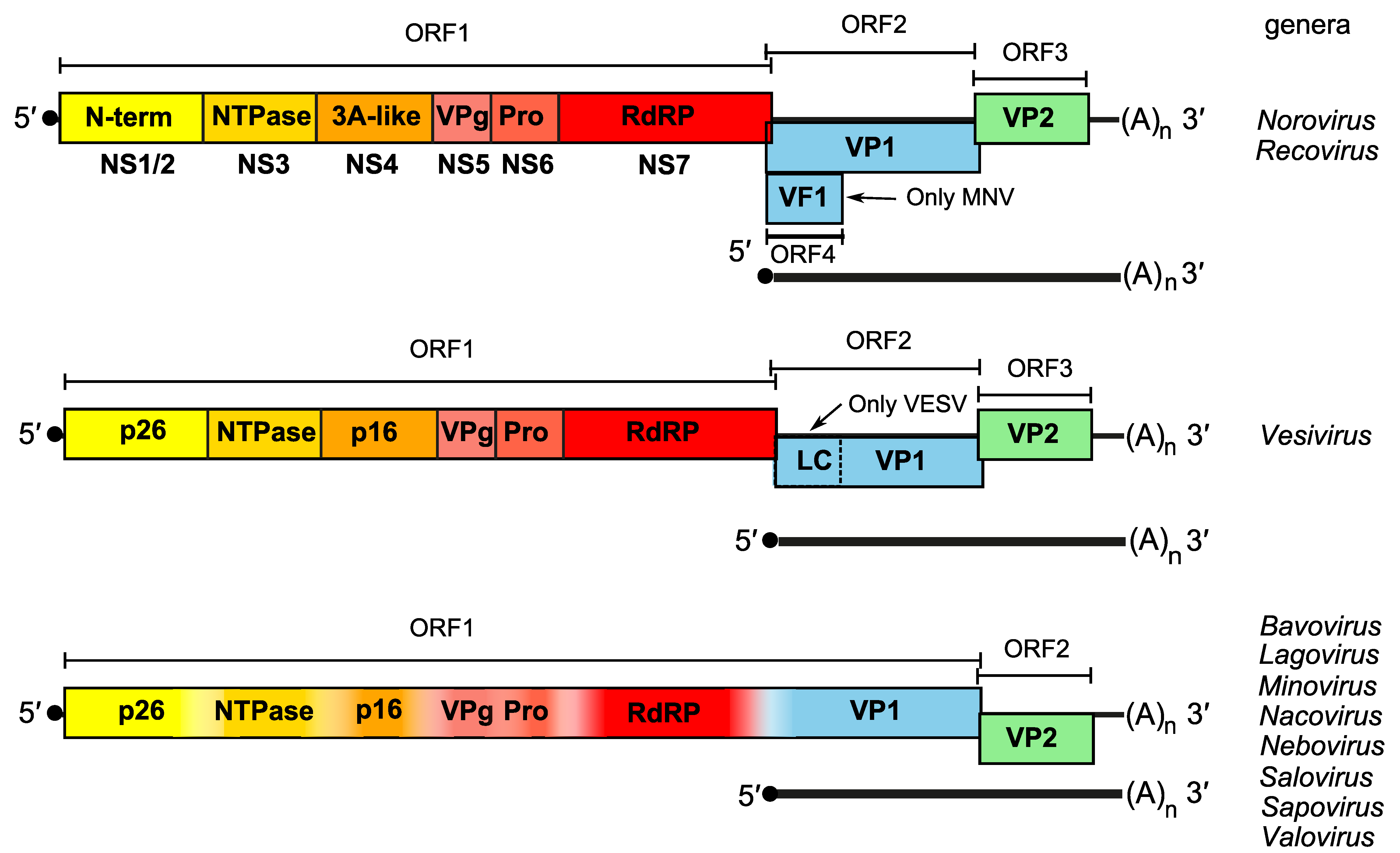 |
| Figure 3. Caliciviridae. Schematic genome organization of members of genera in the family Caliciviridae. Protein VPg is covalently linked to the 5′-ends of both genomic and subgenomic RNAs and is depicted by a black circle. ORF1 encodes a polyprotein that is posttranslationally-cleaved into individual non-structural proteins. ORF2 (and for members of some genera, ORF3 and ORF4) are expressed from subgenomic mRNAs. Cleavage sites have been described for several genera but there is considerable variation in their exact location. Murine norovirus (MNV) is the only norovirus thus far that possesses ORF4 which encodes VF1 at the 5′-end of ORF2, while feline calicivirus (FCV, Vesivirus) encodes a leader capsid (LC). |
Biology
Caliciviruses have been detected from a broad range of mammals as well as from birds and fish. Except for several vesiviruses, individual caliciviruses generally exhibit a natural host restriction and for viruses from genera for which no productive cell culture system is available, this has presented a considerable problem to study their biology.
The marine vesiviruses are closely related genetically and have a broad host range and a wide cell tropism and therefore generally lack host specificity. They have been isolated from swine sea lions, seals, walrus, cetaceans, cattle, rabbits, primates (including man), reptiles and fish.
Transmission of caliciviruses is via direct contact with an infected host or indirectly via contact with faecal material, vomitus or respiratory secretions, contaminated food, water and fomites. In general, no biologic vectors appear to be involved in transmission, however, mechanical, arthropod vector transmission of RHDV and VESV has been described.
Caliciviruses are associated with a number of disease syndromes. VESV produces clinical signs in swine that are indistinguishable from foot-and-mouth disease, signs that include vesicles in the mouth, tongue, lips, snout and feet at the coronary band and between the digits. In addition, VESV may cause encephalitis, myocarditis, fever, diarrhoea, abortion, hepatitis, pneumonia and hemorrhage and failure of recovered animals to thrive. FCV in cats is predominantly associated with oral ulceration and rhinitis, and causes persistent infections. A more severe disease termed virulent-systemic feline calicivirus disease associated with epithelial cytolysis and systemic vascular compromise in susceptible cats, leading to severe oedema and high mortality has been recognized, (Hurley et al., 2004).
In the genus Lagovirus, RHDV is associated with a generalized viraemic infection in which there is massive liver necrosis that triggers a disseminated intravascular coagulation and rapid death in rabbits greater than three months of age (Abrantes et al., 2012). EBHSV is similar to RHDV but appears to be less virulent while non-virulent viruses related to RHDV have also been described.
Human caliciviruses in the genera Norovirus and Sapovirus induce a generally self-limiting acute gastroenteritis (Green 2013, Oka et al., 2015). Vomiting, often with an acute onset, is, together with diarrhea, a consistent and prominent symptom; other symptoms may include nausea, abdominal cramping, fever and malaise. Prolonged disease is seen in immunocompromised individuals. Neboviruses cause diarrhoea and mild upper intestinal lesions in calves (Smiley et al., 2002, Oliver et al., 2006). Viruses from the genus Bavovirus has been associated with enteritis in chickens and viruses from the genus Nacovirus with enteritis and liver enlargements with foci of intrahepatic necrosis and inflammation in chicken and turkeys. Tulane virus is the exemplar isolate of the type species Recovirus A in the genus Recovirus and was originally isolated from a rhesus macaque in a US primate center (Farkas et al., 2008). In rare cases, phylogenetically-distinct recoviruses have been detected in human stool samples in Bangladesh and Venezuela. Valoviruses have been detected in fecal specimens from swine and their disease association is unknown (L'Homme et al., 2009). Minoviruses and Saloviruses cause systemic infections in different fish species (Mor et al., 2017, Mikalsen et al., 2014).
Antigenicity
Division into serotypes has been defined by a lack of cross-neutralization using specific antisera. In the genus Vesivirus, in addition to 13 serotypes of the species Vesicular exanthema of swine virus in pigs [known as vesicular exanthema of swine virus (VESV)], at least 21 additional serotypes have been described from 20 marine and other species (some are known as San Miguel sea lion viruses and other host-specific names, e.g., walrus calicivirus) (Smith et al., 1998). These marine viruses are grouped together phylogenetically as “marine vesiviruses”. Feline calicivirus (FCV) is a member of a different species in the Vesivirus genus and although antigenic variants have been described, FCV represents a single serotype (Neill et al., 1995). A number of other distinct vesiviruses remain to be classified.
Within the lagoviruses, cross-challenge studies in the natural host and experiments with monoclonal antibodies indicate that rabbit hemorrhagic disease virus (RHDV) and European brown hare syndrome virus (EBHSV) are antigenically distinct.
For viruses in the genera Norovirus and Sapovirus antigenic types have been defined by cross-challenge studies, immune-electron microscopy or solid-phase immune-electron microscopy.
For several viruses of the genera Lagovirus, Norovirus and Sapovirus, recombinant virus-like particles (VLPs) have been produced by expression of VP1 in mammalian, insect and plant systems. These VLPs are highly immunogenic and morphologically and antigenically similar to native virions and have become very useful tools to study the structure, antigenicity and immune responses against these viruses. VLPs derived from RHDV are highly immunogenic and vaccinated rabbits survived a lethal dose of RHDV (Guo et al., 2016). Several VLP-based norovirus vaccines that are in clinical trials in humans are well tolerated and show no serious adverse events (Atmar et al., 2011, Kim et al., 2018).
Derivation of names
Calici: from Latin calix, “cup” or “goblet”, from cup-shaped depressions on the virion surface observed by electron microscopy.
Bavo: modified from the type species name Bavaria virus
Lago: from Lagomorpha, the mammalian host order for the prototype strain rabbit hemorrhagic disease virus.
Mino: refers to the fathead minnows fish in which the virus was detected
Naco: refers to novel avian calicivirus
Noro: modified from the type species name Norwalk virus.
Reco: refers to rhesus enteric calicivirus
Salo: refers to Atlantic salmon
Sapo: modified from the type species name Sapporo virus.
Nebo: derived by using the first two letters from Newbury and Nebraska, the first two isolates to be studied together with bo for bovine.
Vesi: from the type species name Vesicular exanthema of swine virus.
Valo: refers to the town St Valérien in Quebec, Canada.
Genus demarcation criteria
Members of different genera in the family Caliciviridae have >60% amino acid sequence difference in the complete VP1 (Table 2).
Table 2. Caliciviridae. Amino acid divergence in VP1 between members of calicivirus genera.
| Genus | Bavovirus | Lagovirus | Minovirus | Nacovirus | Nebovirus | Norovirus | Recovirus | Salovirus | Sapovirus | Valovirus | Vesivirus |
| Bavovirus | 0.040 | ||||||||||
| Lagovirus | 0.815 | 0.168 | |||||||||
| Minovirus | 0.850 | 0.843 | NAa | ||||||||
| Nacovirus | 0.726 | 0.784 | 0.850 | 0.580 | |||||||
| Nebovirus | 0.820 | 0.719 | 0.853 | 0.808 | 0.390 | ||||||
| Norovirus | 0.852 | 0.814 | 0.805 | 0.835 | 0.809 | 0.516 | |||||
| Recovirus | 0.859 | 0.823 | 0.805 | 0.848 | 0.838 | 0.731 | 0.378 | ||||
| Salovirus | 0.860 | 0.829 | 0.775 | 0.847 | 0.822 | 0.784 | 0.814 | 0.170 | |||
| Sapovirus | 0.792 | 0.780 | 0.854 | 0.768 | 0.770 | 0.828 | 0.846 | 0.836 | 0.557 | ||
| Valovirus | 0.857 | 0.846 | 0.823 | 0.833 | 0.841 | 0.714 | 0.648 | 0.808 | 0.847 | 0.020 | |
| Vesivirus | 0.808 | 0.753 | 0.863 | 0.796 | 0.758 | 0.824 | 0.860 | 0.847 | 0.752 | 0.852 | 0.484 |
a only 1 sequence available
Relationships within the family
There are eleven recognized genera within the family Caliciviridae. The phylogenetic relationships are summarized in Figure 4. Caliciviridae.
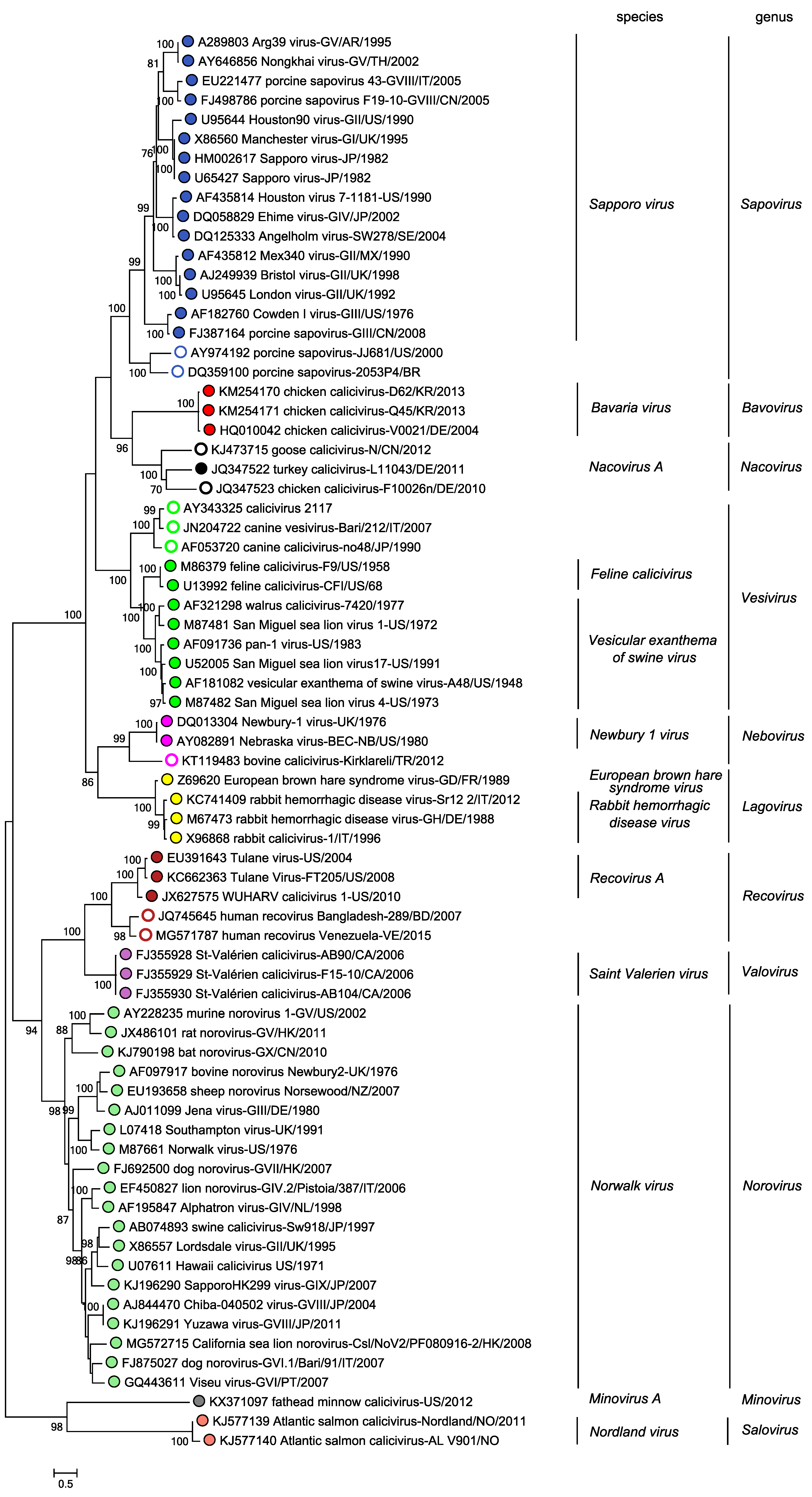 |
| Figure 4. Caliciviridae. Phylogenetic relationships among members of the 11 genera in the family Caliciviridae. Branch tips are labelled with coloured circles to indicate genus assignment, with open circles indicating viruses that have not yet been classified into a species. Maximum-likelihood tree of complete VP1 amino acid sequences of the exemplar and other isolates in each genus. Bootstrap support from 500 replicates is indicated for values > 70%. Multiple sequence alignments were carried out using ClustalW in MEGA X (Kumar et al., 2018). The tree was generated using maximum likelihood method in MEGA7 (Kumar et al., 2016) based on the Le Gascuel 2008 with frequencies model with a gamma distribution of variation including invariant sites. The mid-point rooted tree is drawn to scale with branch lengths measured in the number of substitutions per site. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Caliciviruses have some properties similar to the viruses in the order Picornavirales and the family Potyviridae in that they possess a genome-linked protein, VPg, at the 5′-terminus and a poly(A) tract at the 3′-terminus of the genome. The viral RNA-directed RNA polymerase and the protease of caliciviruses share sequence homology with the picornaviruses.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| avocet calicivirus | MH453804 | |
| bat calicivirus BtCalV-A10 | MH259583 | |
| Beihai conger calicivirus | MG599956 | |
| Beihai fish calicivirus | MG599957 | |
| Beihai rabbitfish calicivirus 1 | MG599959 | |
| Beihai rabbitfish calicivirus 2 | MG599960 | |
| Beihai yellowfin seabream calicivirus | MG599958 | |
| Dongbei arctic lamprey calicivirus 1 | MG599967 | |
| Dongbei arctic lamprey calicivirus 2 | MG599968 | |
| Fujian spotted paddle-tail newt calicivirus | MG599974 | |
| goose calicivirus H146 | KY399947 | |
| Guangdong greater green snake calicivirus | MG599973 | |
| Guangdong pseudohemiculter dispar calicivirus | MG599978 | |
| ruddy turnstone calicivirus | MK189094 | |
| ruddy turnstone calicivirus A | MH453861 | |
| ruddy turnstone calicivirus B | MH453862 | |
| secalivirus (from sewage) | JQ898339 | |
| shelduck calicivirus | MH453874 | |
| Wenling callionymus kaianus calicivirus | MG599966 | |
| Wenling rattails calicivirus 1 | MG599970 | |
| Wenling rattails calicivirus 2 | MG599969 | |
| Wenling rattails calicivirus 3 | MG599971 | |
| Wenling sharpspine skate calicivirus | MG599977 | |
| Wenling yellow goosefish calicivirus | MG599972 | |
| Wuhan carp calicivirus 1 | MG599961 | |
| Wuhan carp calicivirus 2 | MG599963 | |
| Wuhan sharpbelly calicivirus | MG599964 | |
| Wuhan spiny eel calicivirus 1 | MG599962 | |
| Wuhan spiny eel calicivirus 2 | MG599965 | |
| Yancheng osbecks grenadier anchovy calicivirus | MG599979 | |
| Zhejiang gunthers frog calicivirus 1 | MG599975 | |
| Zhejiang gunthers frog calicivirus 2 | MG599976 |
Virus names and virus abbreviations are not official ICTV designations.

