Family: Togaviridae
Rubing Chen, Suchetana Mukhopadhyay, Andres Merits, Bethany Bolling, Farooq Nasar, Lark L. Coffey, Ann Powers and Scott C. Weaver
Corresponding author: Rubing Chen ([email protected])
Edited by: Nick J. Knowles, Stuart G. Siddell and Peter Simmonds
Posted: May 2018, updated April 2019
PDF: ICTV_Togaviridae.pdf
Summary
Togaviridae is a family of small, enveloped viruses with single-stranded positive-sense RNA genomes of 10–12 kb (Table 1. Togaviridae). Within the family, the Alphavirus genus includes a large number of species that are mostly mosquito-borne and pathogenic in their vertebrate hosts. Many are important human and veterinary pathogens (e.g. chikungunya virus, eastern equine encephalitis virus). Before April 2019 the family also contained the genus Rubivirus that has now been moved to the family Matonaviridae.
Table 1. Togaviridae. Characteristics of members of the family Togaviridae.
| Characteristic | Description |
| Typical member | Sindbis virus (J02363), species Alphavirus sindbis |
| Virion | Enveloped, 65–70 nm spherical virions of regular structure with a single capsid protein and 3 envelope glycoproteins |
| Genome | 10–12 kb of positive-sense, non-segmented RNA |
| Replication | Cytoplasmic, in vesicles derived from the plasma membrane/endosomal compartment. Assembled virions bud from plasma membrane |
| Translation | Non-structural proteins are translated from genomic RNA, and structural proteins from subgenomic RNA, both as polyprotein precursors |
| Host range | Humans and nonhuman primates, equids, birds, amphibians, reptiles, rodents, pigs, sea mammals, salmonids, mosquitoes and some other arthropods; most alphaviruses are mosquito-borne |
| Taxonomy | Realm Riboviria; kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Martellivirales; one genus (Alphavirus) including 32 species |
Virion
Morphology
Alphaviruses, members of the only genus in the family Togaviradae, are enveloped viruses. Virions are spherical, enveloped particles about 70 nm in diameter (Figure 1. Togaviridae). The particle consists of a nucleocapsid core surrounded by a lipid bilayer that is embedded with glycoprotein spikes (Cheng et al., 1995, Zhang et al., 2011).
The nucleocapsid core comprises 240 copies of capsid protein that surround the viral genome. The genome is a single-strand of positive-sense RNA that is capped and has a poly-A tail. Some studies have shown host factors are incorporated into the nucleocapsid core, but their quantities are host-dependent (Sokoloski et al., 2013). The lipid bilayer is host-derived from the site of budding. Alphaviruses bud from the plasma membrane although, in arthropods, budding into internal vesicles has also been observed (Gliedman et al., 1975, Lu and Kielian 2000, Strauss and Strauss 1994). Eighty trimeric glycoprotein spikes cover the surface of alphavirions (Cheng et al., 1995, Zhang et al., 2011). Each spike is composed of three E1-E2 heterodimers. However, in some alphaviruses, the E3 glycoprotein remains non-covalently associated to these spikes.
E1 and E2 each contain a single transmembrane domain. E1 has a short cytoplasmic tail that has been shown to be dispensable. E2 has a long (greater than 30 amino acid) cytoplasmic domain that interacts with a hydrophobic pocket in the capsid protein (Lee et al., 1996, Owen and Kuhn 1997, Skoging et al., 1996, Wilkinson et al., 2005, Jose et al., 2012). This interaction between the external glycoprotein spikes and the internal nucleocapsid core is rare in enveloped virions. Both the nucleocapsid core and the trimeric glycoprotein spikes are arranged with T=4 icosahedral symmetry (Zhang et al., 2011). The interaction between the E2 cytoplasmic domain and capsid protein is thought to mediate this organization.
The capsid, E2, and E1 proteins are the minimal proteins required for an infectious virion but the alphaviruses also translate two further proteins, the 6K protein and its frameshifting product, TF (for TransFrame) (Firth et al., 2008). The TF protein is present in virions of at least some species; very rarely is the 6K protein found in the released particle (Snyder et al., 2013, Ramsey and Mukhopadhyay 2017, Ramsey et al., 2017). TF is found in sub-stoichiometric amounts compared to the other structural proteins. Because of its small size and low quantities, its location and organization in the particle is not known.
Physicochemical and physical properties
The sedimentation coefficient of alphavirus particles is 280S. Alphavirus particles have a buoyant density that is between 1.15–1.22 g cm-3 in sucrose gradients (Sokoloski et al., 2013, Kuhn 2013) and between 1.18–1.19 g cm-3 in tartrate gradients (Hernandez et al., 2005). Virions can be physically denatured by treating with various chemicals including urea, formaldehyde, beta-propiolactone, detergents, and acids. Infectivity is also decreased by heat inactivation, by exposure to low pH in the absence of lipid vesicles and exposure to ultraviolet light (Kuhn 2013, Parkman 1965, Park et al., 2016).
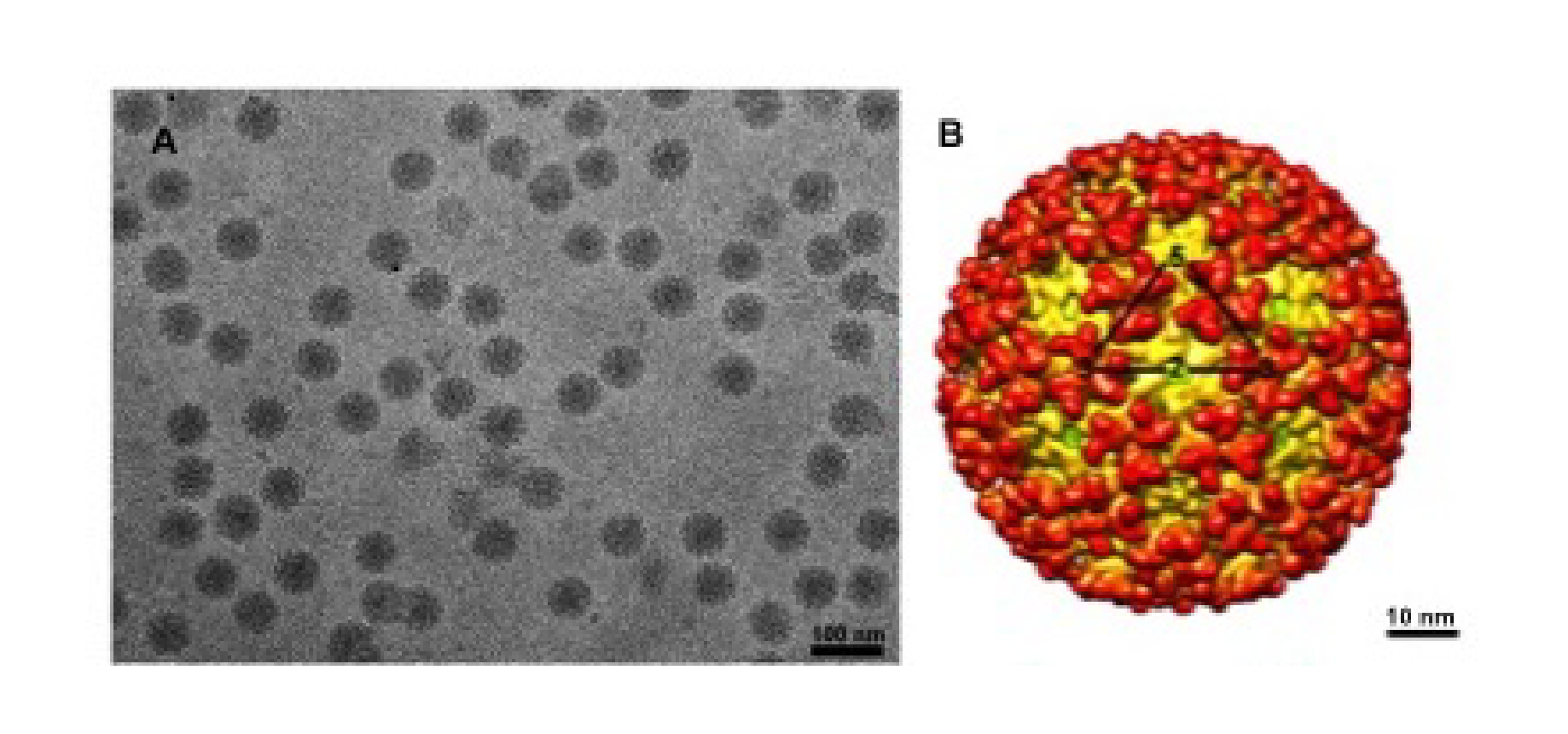 |
| Figure 1. Togaviridae. Images and structures of Togaviridae particles. (A) Chikungunya virus particles purified from BHK cells and flash frozen in vitreous ice (courtesy of J. C-Y Wang and S. Mukhopadhyay). (B) Three dimensional cryo-electron reconstruction of chikungunya virus at 10.2 Å resolution. The triangle outlines one icosahedral unit with the symmetry axes labeled (courtesy of J. C-Y Wang). |
Nucleic acid
Alphaviruses have single-stranded, positive-sense RNA genomes ranging from 9.7–11.8 kb. The genome of alphaviruses consists of a 5ʹ-cap (viral type 0 7meGpppA) and non-coding region followed by genes for the non-structural and structural proteins, a 3ʹ-non-coding region and a poly(A) tail (Strauss and Strauss 1994, Ahola and Kaariainen 1995). The negative-sense RNA of the replication intermediate is neither capped nor contains a poly(A) tail. A subgenomic RNA encoding the structural proteins is transcribed from the negative-sense RNA and consists of a 5′-cap and poly(A) tail.
Proteins
The genome of alphaviruses encodes 10 different proteins. The non-structural proteins (encoded in the order nsP1-nsP4) are important for replicating the viral genome, and the structural proteins (encoded in the order capsid, E3, E2, 6K/TF, and E1) function in virus assembly. The non-structural proteins are not incorporated into virion particles (Strauss and Strauss 1994).
The structural proteins are translated as a polyproteins from the subgenomic mRNA. As soon as capsid is produced it auto-proteolytically cleaves itself from the rest of the polyprotein and forms nucleocapsid cores in the cytoplasm (Choi et al., 1991). E3 contains an ER signal sequence and translocates the polyprotein into the ER. The entire polyprotein crosses the ER membrane (going between lumen and cytoplasm) several times until host proteases cleave it into the individual structural proteins. At a frequency of 10–30%, a ribosomal frameshift event occurs resulting in the production of E3, E2, and TF; E1 is not translated. The host protease signalase cleaves the individual proteins from the polyproteins. The precursor of E3 and E2 proteins (pE2 or p62) forms heterodimers with E1. E1 has been shown to undergo disulphide rearrangement during assembly, and the same is predicted for E2. E3 stabilizes the E2/E1 heterodimers and trimers during spike assembly by preventing dissociation of the complex as it transits through the secretory pathway to the plasma membrane. E3 is released by the host protease furin in the trans-Golgi which converts the spikes to a metastable, fusion competent state, in which form they are transported to the plasma membrane. TF and E2 are palmitoylated. E3, E2, and E1 are glycosylated.
The capsid protein consists of two domains, a highly-charged, N-terminal domain that interacts with the viral RNA in the interior of the nucleocapsid core and a C-terminal domain that has a chymotrypsin-like fold (Choi et al., 1991). The spikes formed by E2 and E1 are responsible for viral entry (Kielian 2014, Kielian et al., 2010, Vaney et al., 2013). The E2 protein binds to the host-cell receptor and interacts with the capsid protein. The E1 protein is a class II fusion protein that mediates fusion between the virus membrane and the host cell membrane in the endosome (Kielian 2014, Kielian et al., 2010, Vaney et al., 2013). There are some reports of fusion occurring at the plasma membrane (Kononchik et al., 2011, Vancini et al., 2015). The atomic structures of E3, E2, and E1 have been determined at both neutral and low pH providing insight to the conformational changes that occur during the entry process (Li et al., 2010, Voss et al., 2010). The exact roles of 6K and TF are not known but 6K has been shown to have viroporin activity (Sanz et al., 1994, Gonzalez and Carrasco 2003) and TF has been shown to be a virulence factor (Snyder et al., 2013).
Lipids
In general the lipid membrane accounts for close to 30% of the virion mass. Alphaviruses contain a host-derived lipid membrane that is acquired during budding from the plasma membrane. In vertebrate cells, alphaviruses bud from regions rich in cholesterol and sphingomyelin but analysis of lipid membrane from purified particles shows a relatively similar lipid composition to that of the plasma membrane (Marquardt et al., 1993, Kielian et al., 2000, Kalvodova et al., 2009).
Carbohydrates
The glycoproteins E3, E2, and E1 have N-linked glycosylation in most alphaviruses (Sefton 1977). However, the locations and numbers of glycosylation sites are not conserved between viruses belonging to different alphavirus species. The carbohydrate moieties incorporated differ depending on the host; virus produced in invertebrates incorporate different carbohydrate residues than virus produced in vertebrate cells. Mutation of glycosylation sites affects virus infectivity and affects the interferon response (Shabman et al., 2008).
Genome organization and replication
Two thirds of the genome of alphaviruses encodes the non-structural polyprotein(s) in a single ORF immediately after a 5′-non-coding region. The proteins are oriented as 5′ – nsP1 (methyltransferase and guanyltransferase) – nsP2 (helicase and protease) – nsP3 (phosphoprotein, (ADP-ribosyl) hydrolase and a key protein for interaction with host factors) – nsP4 (RNA dependent RNA polymerase, terminal adenylyltransferase) – 3′ (Kuhn 2013). There is a stop codon present between the nsP3 and nsP4 genes in the majority of alphaviruses resulting in a limited amount of polyprotein P1234 generated by inefficient read-through. Overlapping with the 3ʹ-end of the non-structural ORF, there is a promoter for transcription of the subgenomic mRNA from which the structural polyprotein is translated (Ou et al., 1982). The structural proteins include the capsid (CP), E3, E2, 6K, TF and E1 proteins. The structural ORF is followed by a non-coding region of varying length (range from 77 to > 980 nucleotides) and finally, a poly(A) tail (Figure 2. Togaviridae).
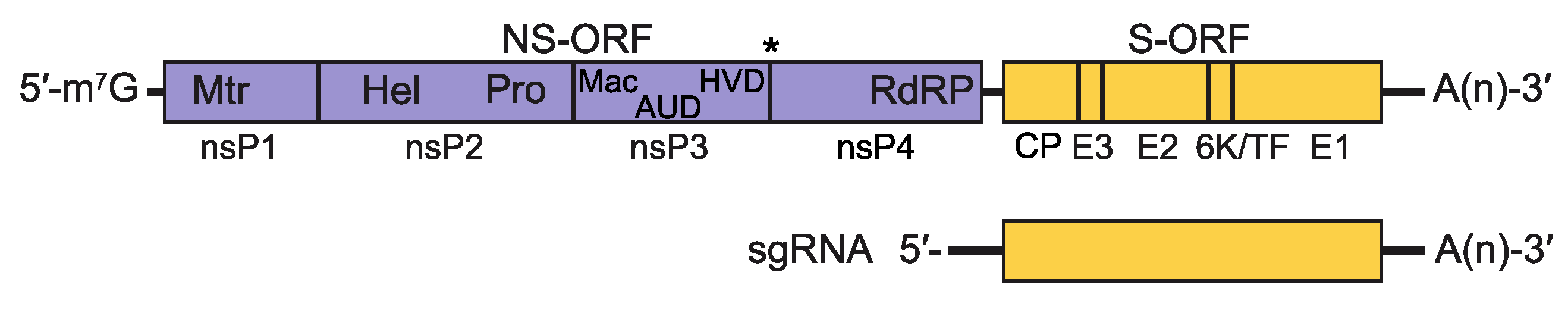 |
| Figure 2. Togaviridae. Alphavirus genomic coding strategy. Shown is genomic RNA with non-coding regions represented as solid black lines and ORFs as open boxes (NS-ORF=non-structural protein ORF; S-ORF=structural protein ORF). Within each ORF, the coding sequences for the proteins processed from the translation product of the ORF are delineated. The asterisk between nsP3 and nsP4 in the NS-ORF indicates the stop codon present in some alphaviruses that must be translationally-read through to produce a precursor containing nsP4. Additionally, within the NS-ORF, the location of motifs associated with the following activities are indicated: (Mtr) methyl transferase, (Pro) protease, (Hel) helicase, (Mac) macro domain, (AUD) alphavirus unique domain, (HVD) hypervariable domain and (RdRP) RNA-dependent RNA polymerase. The sequences encompassed by the sgRNA are also shown. |
Alphavirus replication complexes (termed spherules) are initially formed at the plasma membrane of infected vertebrate cells (Frolova et al., 2010, Spuul et al., 2010). The initial P1234 polyprotein is cleaved by the viral protease activity of the nsP2 protein into P123 precursor protein and nsP4; these proteins form the replication complex associated with negative-sense RNA replication (Lemm et al., 1994). This negative-sense RNA forms a duplex with the positive-sense (genomic) strand and serves as the template in the synthesis of a full-length, positive-sense RNA that will eventually be encapsidated, as well as a subgenomic 26S mRNA that encodes the viral structural proteins (Pietilä et al., 2017) (Figure 3. Togaviridae). Cleavage of the P123 polyprotein is highly regulated and generates nsP1, nsP2 and nsP3 individual proteins, which are involved in the synthesis of positive-sense RNA (Lemm et al., 1994, Vasiljeva et al., 2003). Both alphavirus non-structural proteins and RNAs interact with multiple cellular proteins; some of these interactions are essential for replication (Sokoloski et al., 2010, Kim et al., 2016).
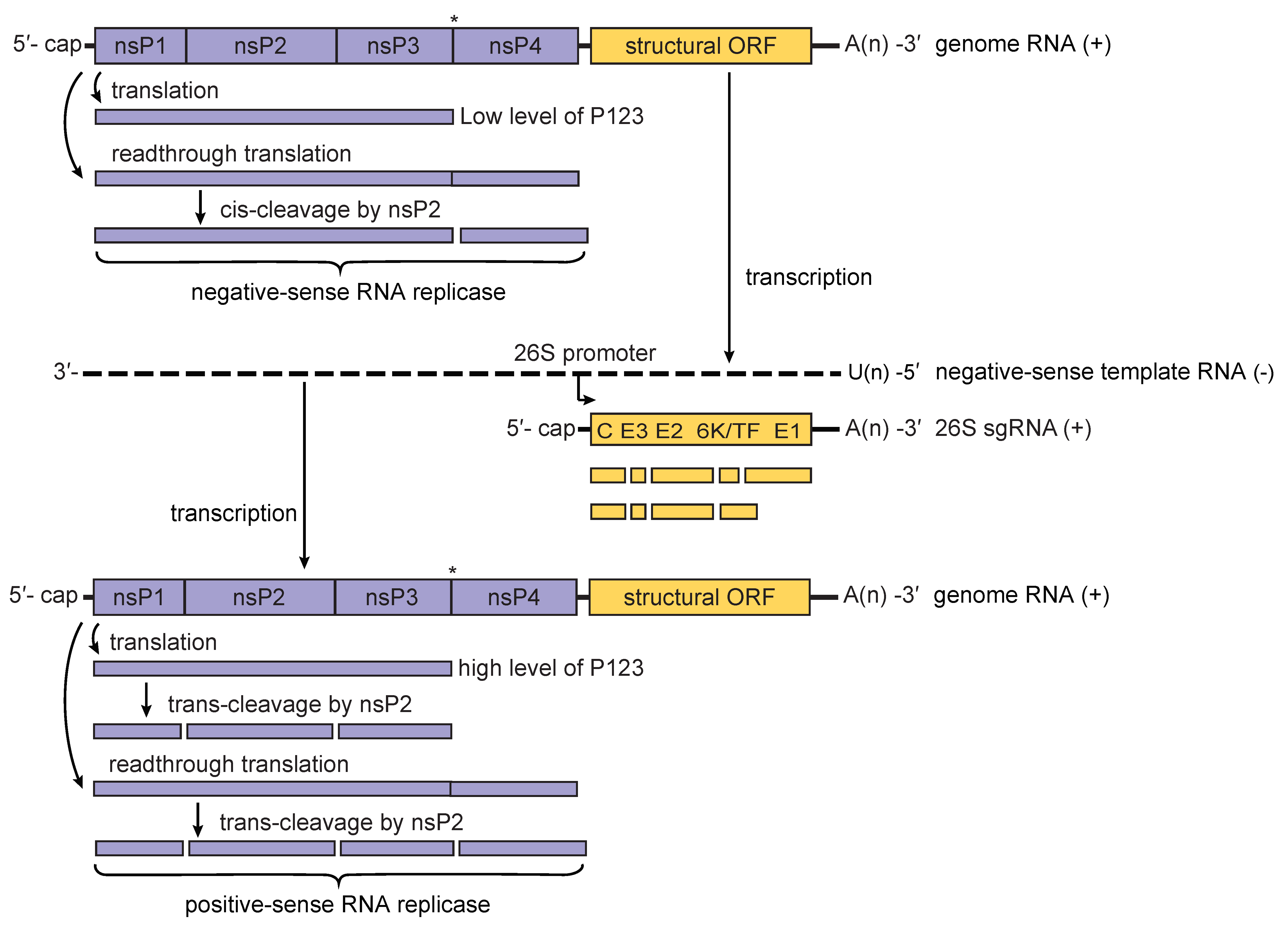 |
| Figure 3. Togaviridae. Model for the processing of the alphavirus nonstructural polyprotein during replication. When low levels of P123 are present, cis-cleavage of P1234 generates the negative-sense RNA replicase of the virus. This results in primarily negative-sense RNA being transcribed from the incoming genomic RNA of the virus (upper panel). Synthesis of negative-sense strand RNA is followed by cis-cleavage of P123 into nsP1 and P23; the latter is cleaved in trans into nsP2 and nsP3. This results in a shift from the production of primarily negative-sense RNA to primarily positive-sense RNA. As the level of the trans-acting nsP2 protease rises in the infected cell, cleavage of P1234 in trans generates P12 and P34 polyproteins that are unable to form RNA replicase complexes. Eventually, replicase complexes capable of producing negative-sense RNA will no longer be present in the infected cell resulting in the complete cessation of negative-sense RNA synthesis (lower panel). The presence of a leaky opal termination codon (indicated by an asterisk) in the virus genome is believed to lead to a more rapid conversion to the production of positive-sense RNA. |
Alphavirus structural polyproteins are translated from a subgenomic mRNA. The predominant translation product is CP/E3/E2/6K/E1, but, at a low frequency, there is a (-1) translational frameshifting event that produces CP/E3/E2/TF. Alphavirus polyproteins are then cleaved by both viral and cellular proteases to produce individual structural proteins (Firth et al., 2008). The glycoproteins that are produced are inserted into the endoplasmic reticulum during translation and are translocated to the plasma membrane. Upon generation of a sufficient amount of CP protein, this protein assembles with the viral RNA to form the viral nucleocapsids in the cytosol. Budding through the plasma membrane leads to the acquisition of a lipid envelope containing the two main membrane glycoproteins E1 and E2 (Frey 1994, Jose et al., 2009).
Biology
Host Range
The genus Alphavirus mainly consists of mosquito-borne viruses although other hematophagous insects, including ticks, lice, and cliff swallow bugs, have been implicated in transmission (Lwande et al., 2013, Hayes et al., 1977). Vertebrate hosts include humans, non-human primates, equids, birds, amphibians, reptiles, rodents, and pigs (Kuhn 2013). There are two aquatic alphaviruses, southern elephant seal virus and salmon pancreas disease virus, infecting sea mammals and fish respectively. Furthermore, representing a new host-restricted complex within the alphaviruses, Eilat virus (EILV), has been shown to only infect insect cells and is incapable of replicating in vertebrate cells (Nasar et al., 2012).
Transmission
With a few exceptions, most alphaviruses are transmitted alternately between their vertebrate and invertebrate hosts. For many alphaviruses, humans are dead-end hosts, incapable of developing sufficient viremia to infect mosquitoes, although human-mosquito-human transmission has been implicated in chikungunya virus (CHIKV) outbreaks. The aquatic alphaviruses, which include southern elephant seal virus (La Linn et al., 2001) and salmon pancreas disease virus (Weston et al., 1999), appear to be transmitted horizontally, although both viruses have been isolated from lice, suggesting arthropod-borne transmission may also play a role. The insect-specific Eilat virus is defective for replication in vertebrates, and therefore is probably maintained via vertical transmission in mosquitoes (Nasar et al., 2012).
Geographical Distribution
The alphaviruses have a worldwide distribution, inhabiting all continents except Antarctica (Powers et al., 2001). The alphaviruses are geographically restricted based on preferred ecological conditions, reservoir hosts and vector species, but continue to move around the globe and colonize new areas. Based on their distribution, alphaviruses have classically been described as Old World or New World viruses. However, CHIKV, belonging to the Old World Semliki Forest complex, was introduced in 2013 from the South Pacific via the Caribbean to North and South America, and in 2014 from Angola to Brazil, in both cases causing severe outbreaks and local transmission (Leparc-Goffart et al., 2014, Nunes et al., 2015).
Pathogenicity
Most alphaviruses are cytopathic to vertebrate cells and cause a short febrile illness that can lead to prolonged arthritis or encephalitis, but are rarely fatal.
Antigenicity
The alphaviruses were originally described as Group A arboviruses based upon their antigenic cross-relationships. Using specific serological testing, antigenic complexes were proposed where all members of a particular complex were closely related to each other. Eight (eleven including the fish, seal and mosquito-specific alphaviruses) such complexes are described whose members, for the most part, are also genetically clustered (Kuhn 2013).
Derivation of names
Alpha: from Greek letter α., originally group A arboviruses.
Aura virus: name of small river in Belém, Brazil where the virus was originally isolated (Causey et al., 1963).
chikungunya virus: chikungunya derives from a word in the Kimakonde/Makonde language, meaning “to become contorted”, and describes the stooped appearance of sufferers with joint pain (arthralgia).
getah virus: isolated near rubber plantations; Getah means rubber in Malay.
Madariaga virus: first isolates from General Madariaga Partido, Buenos Aires Province, Argentina.
Mosso das Pedras virus: named after isolation locality in Brazil (Calisher et al., 1982).
onyong-nyong virus: onyong-nyong means "weakening of the joints" in the Nilotic language
Ross River virus: isolated near Ross River in Australia
Semliki Forest virus: first isolated from Semliki Forest in Uganda
Sindbis virus: isolated near Sindbis, Egypt
Toga: from Latin toga, meaning “cloak”
Trocara virus: after Trocará, Brazil.
Una virus: name of small river in Belém, Brazil where the virus was originally isolated (Causey et al., 1963).
Species demarcation criteria
Species demarcation criteria in the genus include:
- Nucleotide and deduced amino acid sequences.
- Antigenic characteristics.
- Vector association.
- Host association.
- Disease association.
- Ecological characteristics.
Species demarcation considers a combination of each of the criteria listed above. Whereas members of most species show at least 10% difference in amino acid sequence identity over entire coding regions, there is not a clear cutoff of sequence divergence to provide absolute species demarcation. For example, for Venezuelan equine encephalitis virus (VEEV), there are multiple subtypes with amino acid identities as low as 85%. However, although Everglades, Tonate, and Mucambo viruses are phylogenetically located within the VEEV cluster (Figure 4. Togaviridae) and have 94–97% amino acid identity with each other or the closest related VEEV strains, they are considered to belong to different individual species due to differences in several of the traits listed above. For example, the justification for Everglades virus remaining as a distinct species is based upon its avirulence for equids, as well as a different rodent host and mosquito vector usage in Southern Florida.
The reason that some very divergent strains such as those of VEEV and Sindbis virus are considered subtypes but not species reflects the lack of phenotypic information. Madariaga virus, previously considered to comprise 3 subtypes within the species Eastern equine encephalitis virus, was assigned to the species Madariaga virus based on major differences in vector usage and human virulence for most strains (Arrigo et al., 2010).
Relationships within the family
Among alphaviruses, salmon pancreatic disease virus (SPDV) is the most divergent, with sequence similarity only in parts of the structural and non-structural proteins. However, this may not necessarily mean that this virus is ancestral, but could instead reflect its adaptation to the fish host and a lack of evolutionary constraints for other alphaviruses associated with alternating transmission between a mammalian host and arthropod vector.
An alignment of alphavirus sequences (excluding SPDV), demonstrates a high level of heterogeneity in the hypervariable region (HVR) of the nsP3 gene, the capsid gene, and a few short regions scattered throughout the genome where accurate alignment cannot be made. In addition, a recombination event was involved in the origination of western equine encephalitis, Highlands J and Fort Morgan viruses, where the parental sequences were derived from both the eastern equine encephalitis virus complex (donating non-structural proteins genes and capsid genes) and the Whataroa/Sindbis lineage ancestor (donating the envelope protein genes). A phylogenetic tree based on the conserved regions of envelope genes is shown in Figure 4. Togaviridae.
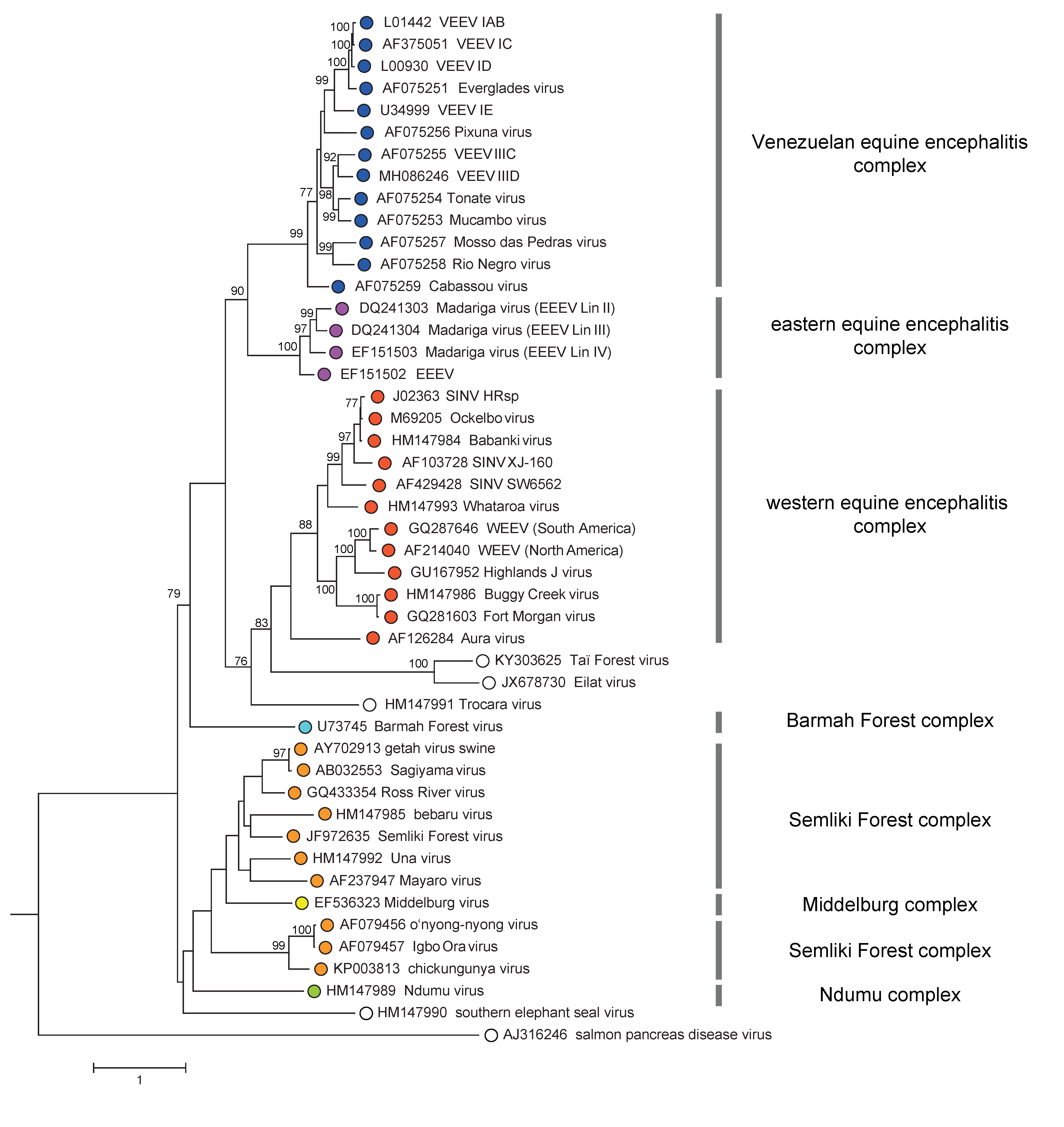 |
| Figure 4. Togaviridae. Phylogenetic tree of representative isolates of all alphavirus species generated from a conserved region of envelope protein gene nucleotide sequences (2184 nt) using the GTR+I+Γ substitution model and Maximum likelihood method. The tree is mid-point rooted. Bootstrap values above 70 generated by 1000 replicates of neighbour-joining tree are indicated next to the main branches. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
There are 3 major clades in the Alphavirus genus tree, all supported by high bootstrap values. The first major clade further diverges into Venezuelan equine encephalitis and eastern equine encephalitis complexes. The second bifurcates into Trocara virus, Eilat virus complex, and western equine encephalitis complex, sequentially. The last one contains Barmah Forest virus, salmon pancreas disease virus, southern elephant seal virus, Ndumu virus, Middleburg virus, and Semliki Forest virus complex. Notably, despite Middleburg virus being phylogenetically located within the Semliki Forest complex, it is considered a distinct antigenic complex.
Relationships with other taxa
Flaviviruses were previously included in the family Togaviridae due to some biological and virological similarities to alphaviruses. An atomic resolution crystal structure of an alphavirus E1 protein shows a folding pattern related to the E protein of flaviviruses, suggesting homology of at least some genes between these families (Jose et al., 2009). However, their distinct genetic organization and mode of replication led to the eventual separation of these groups.
Until 2019 the family Togaviridae also contained the genus Rubivirus. This classification was based on similarities in virion structure and gene expression strategy. Limited sequence similarity between conserved regions of the alphavirus genome and their counterparts in the rubivirus genome has also been reported. However, rubiviruses differ from alphaviruses in their mode of transmission, genome organization, virion structure and sequence relationships, and so have been moved to the family Matonaviridae.
Similarities in replication proteins among members of RNA viruses from several plant and insect infecting virus families, and with members of the family Hepeviridae provide support for an alphavirus-like superfamily (Rozanov et al., 1992).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Taï Forest alphavirus | KY303625 | TALV |
Virus names and virus abbreviations are not official ICTV designations.

